Nomenclature
Short Name:
LRRK2
Full Name:
Leucine-rich repeat kinase 2
Alias:
- Dardarin
- DKFZp434H2111
- RIPK7
- ROCO2
- EC 2.7.11.1
- FLJ45829
- Leucine-rich repeat kinase 2
- PARK8
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
LRRK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
286103
# Amino Acids:
2527
# mRNA Isoforms:
1
mRNA Isoforms:
286,103 Da (2527 AA; Q5S007)
4D Structure:
Interacts with PARK2
1D Structure:
Subfamily Alignment
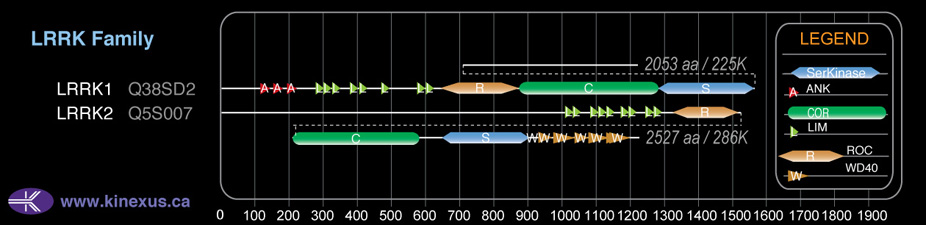
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S3, S5, S633, S634, S784, S788, S837, S850, S858, S860, S865, S895, S898, S908, S910, S912, S926, S933, S935, S954, S955, S958, S962, S971, S973, S975, S976, S979, S1025, S1058, S1124, S1157, S1159, S1219, S1228, S1253, S1283, S1292, S1345, S1403, S1443, S1444, S1445, S1457, S1467, S1508, S1536, S1647, S1853, S1913, S2032+, S2166, S2257.
Threonine phosphorylated:
T424, T489, T496, T524, T776, T826, T833, T838, T1024, T1176, T1343, T1348, T1349, T1357, T1368, T1404, T1410+, T1452, T1470, T1491, T1503, T1612, T1849, T1912, T1967, T1969, T2031, T2035+, T2237, T2460, T2483, T2524, .
Tyrosine phosphorylated:
Y636, Y707, Y1332, Y1402, Y1485, Y2023, Y2449.
Ubiquitinated:
K1118, K1129.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 96
96
1361
9
1040
 30
30
428
3
370
 -
-
-
-
-
 12
12
176
37
157
 46
46
654
10
439
 3
3
43
9
17
 0.9
0.9
13
10
8
 60
60
852
12
1849
 -
-
-
-
-
 23
23
328
30
136
 6
6
91
6
78
 100
100
1413
12
742
 13
13
182
2
129
 28
28
401
2
402
 12
12
173
6
80
 4
4
51
7
31
 4
4
61
69
48
 36
36
508
2
3
 16
16
233
20
607
 44
44
621
30
369
 27
27
388
6
222
 34
34
480
4
139
 -
-
-
-
-
 7
7
98
4
58
 12
12
171
6
74
 52
52
731
25
868
 22
22
313
2
117
 34
34
487
2
61
 55
55
771
2
481
 5
5
77
14
47
 -
-
-
-
-
 4
4
60
13
62
 26
26
369
51
659
 56
56
791
26
622
 1.3
1.3
19
22
14
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.1
99.1
99.5
99 -
-
-
95.5 -
-
-
91 -
-
-
- 90.1
90.1
94
92 -
-
-
- 86.6
86.6
93.1
87 86.5
86.5
93.4
87 -
-
-
- -
-
-
- -
-
-
73 -
-
-
63.5 42.6
42.6
59
50 -
-
-
- -
-
-
- 20.4
20.4
38.5
- -
-
-
- 29.3
29.3
49.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
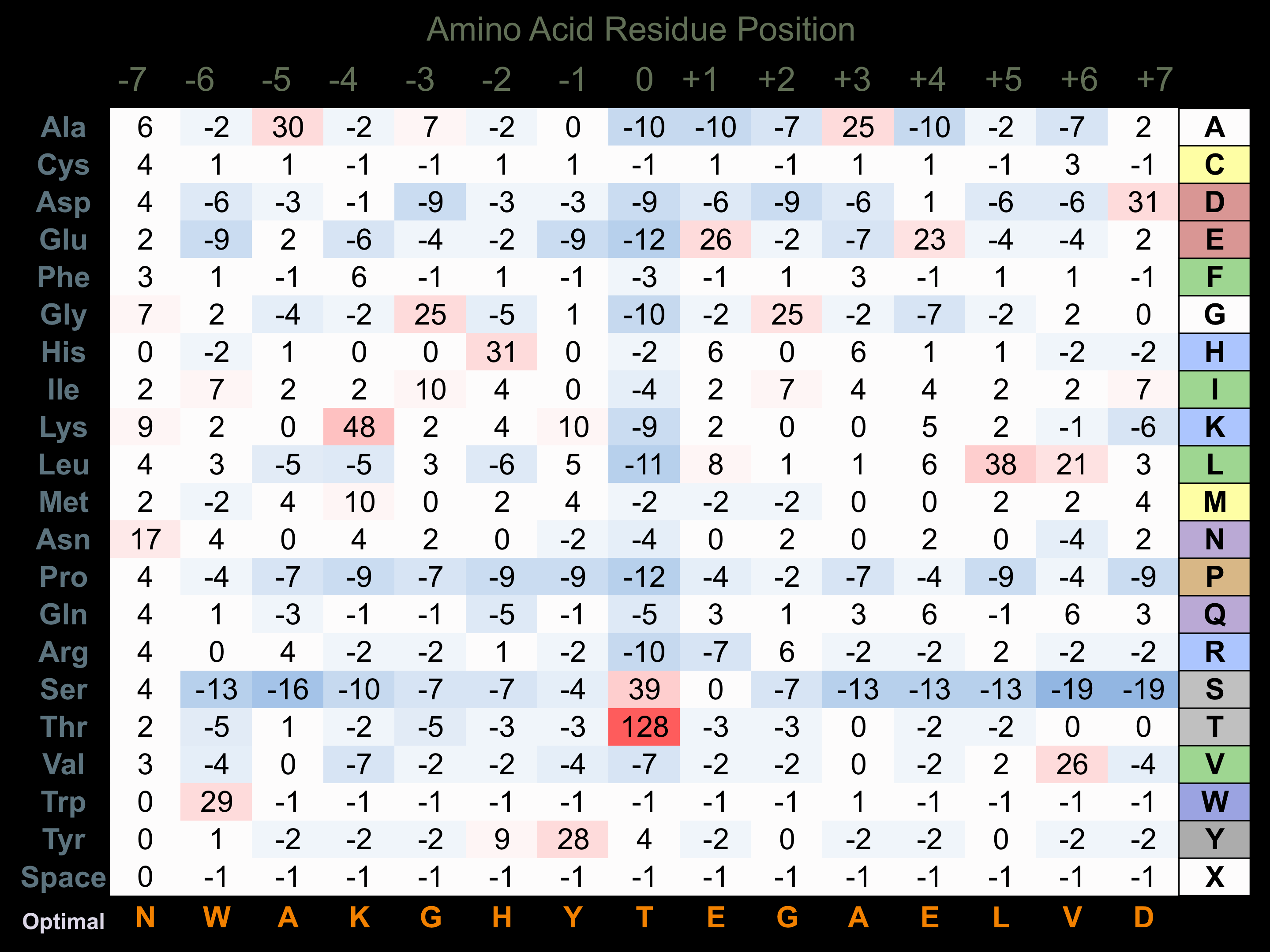
Matrix Type:
Experimentally derived from alignment of 61 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Parkinson's disease (PD); LRRK2-related Parkinson's disease (PD); Tremor; Essential tremor; Parkinson's disease 8; Primary progressive aphasia; Rheumatoid arthritis; Movement disease; Parkinson's disease 1
Comments:
(PD) is a neurodegenerative movement disorder, characterized by the degeneration of the dopaminergic neurons in the substantia nigra of the midbrain. Synptoms of PD include trembling of hands, arms, legs, and face, stiffness in the arms and legs, bradykinesia, and poor coordination and balance, as well as the presence in some patients of neurofibrillary MAPT (tau)-positive and Lewy bodies. LRRK2 (leucine-rich repeat serine/threonine-protein kinase 2) functions as a positive regulator of autophagy via a calcium-dependent activation of the CaMKK/AMPK signalling pathway. Additionally, in combination with RAB29, LRRK2 functions in the retrograde trafficking pathway for the recycling of proteins between lysosomes and the golgi apparatus. LRRK2 also regulates the morphology of neuronal processes in the central nervous system. The LRRK2 protein contains a leucine-rich repeat domain, a Roc domain, a COR domain, a MAPKKK domain, and a WD40 domain. Mutations in the LRRK2 gene are one of the most common genetic causes of inherited Parkinson's disease. In the brain, highest expression of the LRRK2 gene is seen in the putamen and substantia nigra, most notable as a region with high density of dopaminergic neurons. Thus it has been suggested that loss of LRRK2 expression in these regions may result in the loss of trophic support to dopamine neurons, contributing to the disease pathology of PD. In rat hippocampal neurons, LRRK2 has been shown to co-localize with alpha/beta-tubulin, which when polymerized into microtubules are critically involved in the pathogenesis of neurodegenerative diseases, such as PD. In addition, mutant forms of LRRK2 have been associated with substantial increases in tubulin phosphorylation, which stabilizes the microtubules. This is suggested as a potential contributor to the pathology of PD, as it may disrupt neurite outgrowth, axonal transport, and synapse formation. LRRK2 has also been demonstrated to interact directly with PRDX3, an important mitochondrial antioxidant. Several mutations in the LRRK2 gene are associated with increased phosphorylation of the PRDX3 protein, which results in decreased anti-oxidant activity, mitochondrial dysfunction, increased oxidative damage, and cell death. This finding suggests a link between compromised anti-oxidant function, induced by LRRK2, and neurodegenerative diseases (e.g. PD). Overall, mutation of the LRRK2 protein is associated with the increased phosphorylation of several downstream gene targets and linked with the development of PD.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Classical Hodgkin lymphomas (%CFC= -46, p<0.002); Clear cell renal cell carcinomas (cRCC) (%CFC= +104, p<0.004); Ovary adenocarcinomas (%CFC= -60, p<0.048); Papillary thyroid carcinomas (PTC) (%CFC= +45, p<0.0001); and Vulvar intraepithelial neoplasia (%CFC= -47, p<0.008). The COSMIC website notes an up-regulated expression score for LRRK2 in diverse human cancers of 344, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25124 diverse cancer specimens. This rate is only 26 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.13 % in 1270 large intestine cancers tested; 0.1 % in 864 skin cancers tested; 0.09 % in 603 endometrium cancers tested; 0.06 % in 589 stomach cancers tested; 0.05 % in 833 ovary cancers tested; 0.04 % in 1364 kidney cancers tested; 0.03 % in 710 oesophagus cancers tested; 0.03 % in 548 urinary tract cancers tested; 0.03 % in 238 bone cancers tested; 0.03 % in 1824 lung cancers tested; 0.02 % in 1512 liver cancers tested; 0.02 % in 1316 breast cancers tested; 0.01 % in 273 cervix cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested; 0.01 % in 1459 pancreas cancers tested; 0.01 % in 1062 upper aerodigestive tract cancers tested; 0 % in 881 prostate cancers tested; 0 % in 2082 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: I1294V (9).
Comments:
Fifteen deletions, 7 insertions and 3 complex mutations are noted on the COSMIC website.

