Nomenclature
Short Name:
BRSK2
Full Name:
BR serine-threonine-protein kinase 2
Alias:
- BR serine/threonine kinase 2
- C11orf7
- Serine/threonine kinase 29
- Serine/threonine- protein kinase 29
- STK29
- EC 2.7.11.1
- PEN11B
- SAD1B
- Serine,threonine-protein kinase 29
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
BRSK
Specific Links
Structure
Mol. Mass (Da):
81,633
# Amino Acids:
736
# mRNA Isoforms:
6
mRNA Isoforms:
84,832 Da (766 AA; Q8IWQ3-5); 81,633 Da (736 AA; Q8IWQ3); 77,598 Da (696 AA; Q8IWQ3-4); 75,149 Da (674 AA; Q8IWQ3-2); 74,741 Da (668 AA; Q8IWQ3-3); 68,740 Da (614 AA; Q8IWQ3-6)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 19 | 270 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
R437.
Serine phosphorylated:
S294, S367, S382, S393, S410, S412, S417, S423, S426, S427, S435, S439, S466, S481, S489, S506, S512, S513, S520.
Threonine phosphorylated:
T174+, T260+, T431, T443, T463, T509.
Tyrosine phosphorylated:
Y15.
Ubiquitinated:
K48.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 51
51
465
22
656
 1.2
1.2
11
12
9
 0.6
0.6
5
1
0
 35
35
318
73
708
 26
26
235
27
223
 22
22
200
54
553
 19
19
169
32
436
 92
92
833
22
1227
 20
20
181
13
219
 8
8
73
71
196
 14
14
131
17
328
 47
47
428
104
471
 0.2
0.2
2
12
1
 1.1
1.1
10
9
5
 54
54
487
14
1196
 1.3
1.3
12
12
7
 3
3
23
105
118
 1.4
1.4
13
9
6
 2
2
22
64
17
 27
27
249
87
248
 35
35
320
13
733
 14
14
126
15
312
 0.3
0.3
3
2
1
 3
3
28
9
24
 14
14
129
13
293
 100
100
908
44
1809
 0.9
0.9
8
18
8
 1.4
1.4
13
9
6
 2
2
15
9
9
 12
12
105
28
116
 41
41
371
18
270
 36
36
326
26
423
 3
3
26
69
128
 70
70
640
52
567
 15
15
132
35
219
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
92 -
-
-
- -
-
-
97.5 -
-
-
- 89.8
89.8
90.3
99 -
-
-
- 97.9
97.9
98.7
98 25.4
25.4
43.2
98 -
-
-
- -
-
-
- 27.4
27.4
43.9
93 -
-
-
86 25.1
25.1
37.9
88.5 -
-
-
- 50.9
50.9
63.5
63 -
-
-
- 23.1
23.1
37.3
65 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 27.8
27.8
43.2
- 22.9
22.9
33.9
39 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC25B - P30305 |
| 2 | CDC25C - P30307 |
| 3 | WEE1 - P30291 |
| 4 | NUAK1 - O60285 |
Regulation
Activation:
Activated by phosphorylation on Thr-174 by STK11 in complex with STE20-related adapter-alpha (STRAD alpha) pseudo kinase and CAB39.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
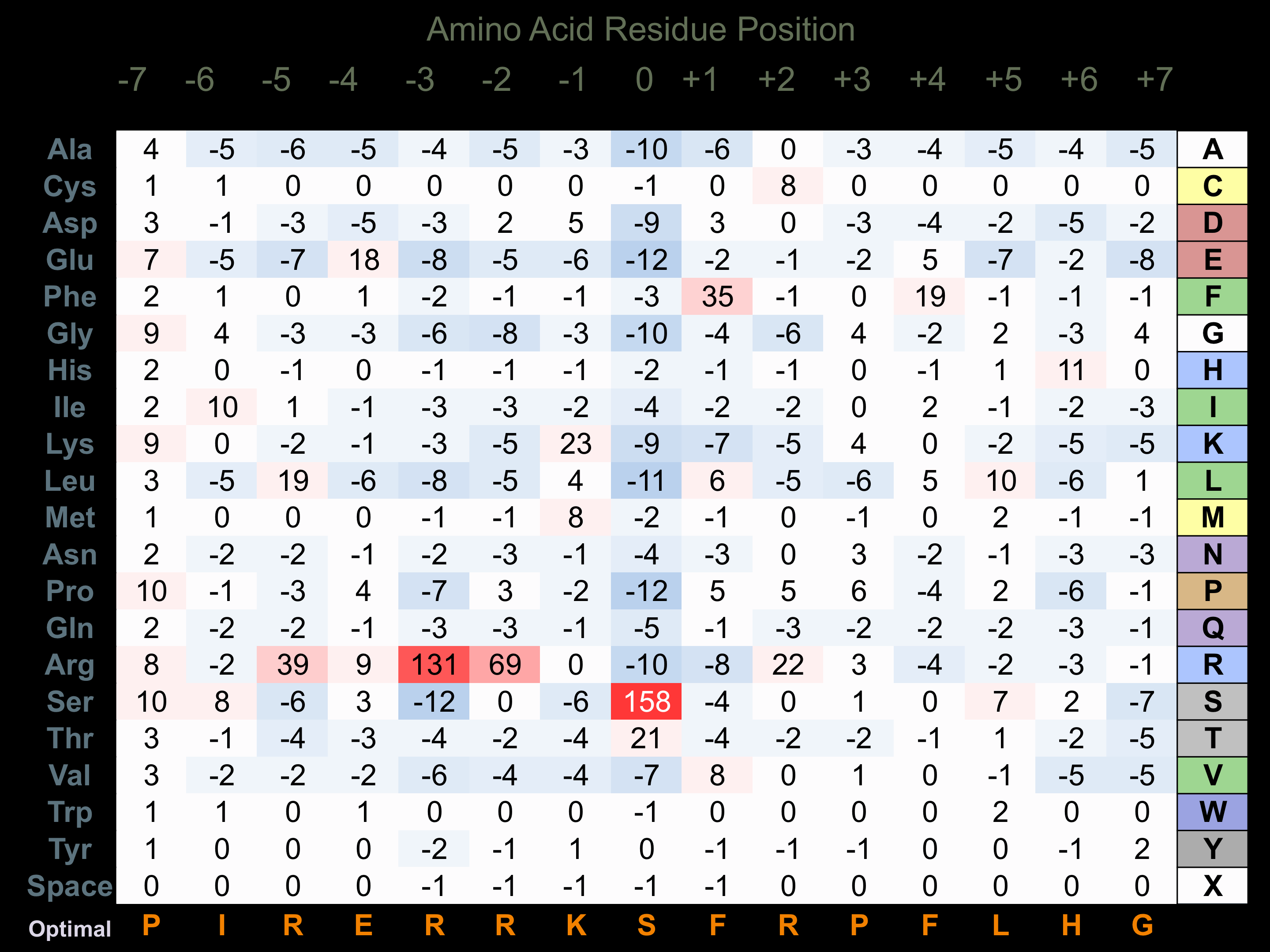
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Limbic encephalitis
Comments:
Mutations on multiple sites can be either activatory or inhibitory on BRSK2's phosphotransferase activity. Example of diseases that may be associated with BRSK2 are limbic encephalitis and small-cell lung cancer (SCLC). BRSK2 may be an autoantigen involved in the pathogenesis of SCLC-associated limbic encephalitis and SCLC patients with paraneoplastic neurological syndromes have been reported to have antibodies against BRSK2 (PubMed:16165222).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24434 diverse cancer specimens. This rate is -40 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.31 % in 864 skin cancers tested; 0.25 % in 273 cervix cancers tested; 0.14 % in 1229 large intestine cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,009 cancer specimens
Comments:
Only 3 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

