Nomenclature
Short Name:
LMR2
Full Name:
KPI-2 protein
Alias:
- 2900041G10Rik
- AATYK2
- KIAA1079
- Kinase LMR2
- Kinase phosphatase inhibitor 2
- LMTK2
- BREK
- CDK5,p35-regulated kinase
- Cprk
- EC 2.7.11.1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Lmr
SubFamily:
NA
Structure
Mol. Mass (Da):
164900
# Amino Acids:
1503
# mRNA Isoforms:
1
mRNA Isoforms:
164,900 Da (1503 AA; Q8IWU2)
4D Structure:
Interacts with PPP1C and inhibitor-2.
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 137 | 409 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S473, S525, S587, S630, S788, S883, S886, S955, S1012, S1107, S1111, S1117, S1124, S1126, S1248, S1251, S1291, S1294, S1397, S1446, S1450+, S1465, S1496, S1497.
Threonine phosphorylated:
T90, T582, T602, T805, T876, T996, T1088, T1123, T1445.
Tyrosine phosphorylated:
Y476, Y500, Y1100, Y1448, Y1468.
Ubiquitinated:
K150, K430.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 13
13
914
31
1052
 1
1
94
12
182
 2
2
111
15
111
 6
6
409
123
1837
 8
8
530
31
528
 0.3
0.3
18
55
21
 5
5
340
43
633
 14
14
949
45
3152
 3
3
194
10
192
 4
4
256
123
558
 2
2
119
34
225
 6
6
421
148
494
 0.7
0.7
50
26
52
 2
2
127
10
172
 2
2
161
33
216
 1
1
66
19
91
 5
5
310
273
1552
 2
2
162
23
235
 0.9
0.9
60
99
81
 7
7
463
112
473
 1
1
90
31
95
 2
2
103
34
133
 2
2
116
16
79
 2
2
107
24
122
 2
2
110
32
131
 14
14
925
81
2354
 1
1
70
29
104
 2
2
162
23
315
 1
1
66
23
67
 1
1
66
42
53
 10
10
661
18
453
 100
100
6841
35
11268
 1
1
79
76
194
 10
10
668
78
611
 2
2
154
48
431
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.3
99.3
99.7
99 31
31
46.2
- -
-
-
77 -
-
-
81 78.1
78.1
83.7
84 -
-
-
- 76.1
76.1
84
78 72.6
72.6
80.4
74 -
-
-
- 47.1
47.1
56.5
- 54.8
54.8
68.6
59 -
-
-
58 44.2
44.2
59.6
- -
-
-
- -
-
-
- 20.6
20.6
39.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PPP1R2 - P41236 |
| 2 | PPP1R2P3 - Q6NXS1 |
| 3 | CDK5 - Q00535 |
| 4 | CDK5R1 - Q15078 |
| 5 | PPP1CA - P62136 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
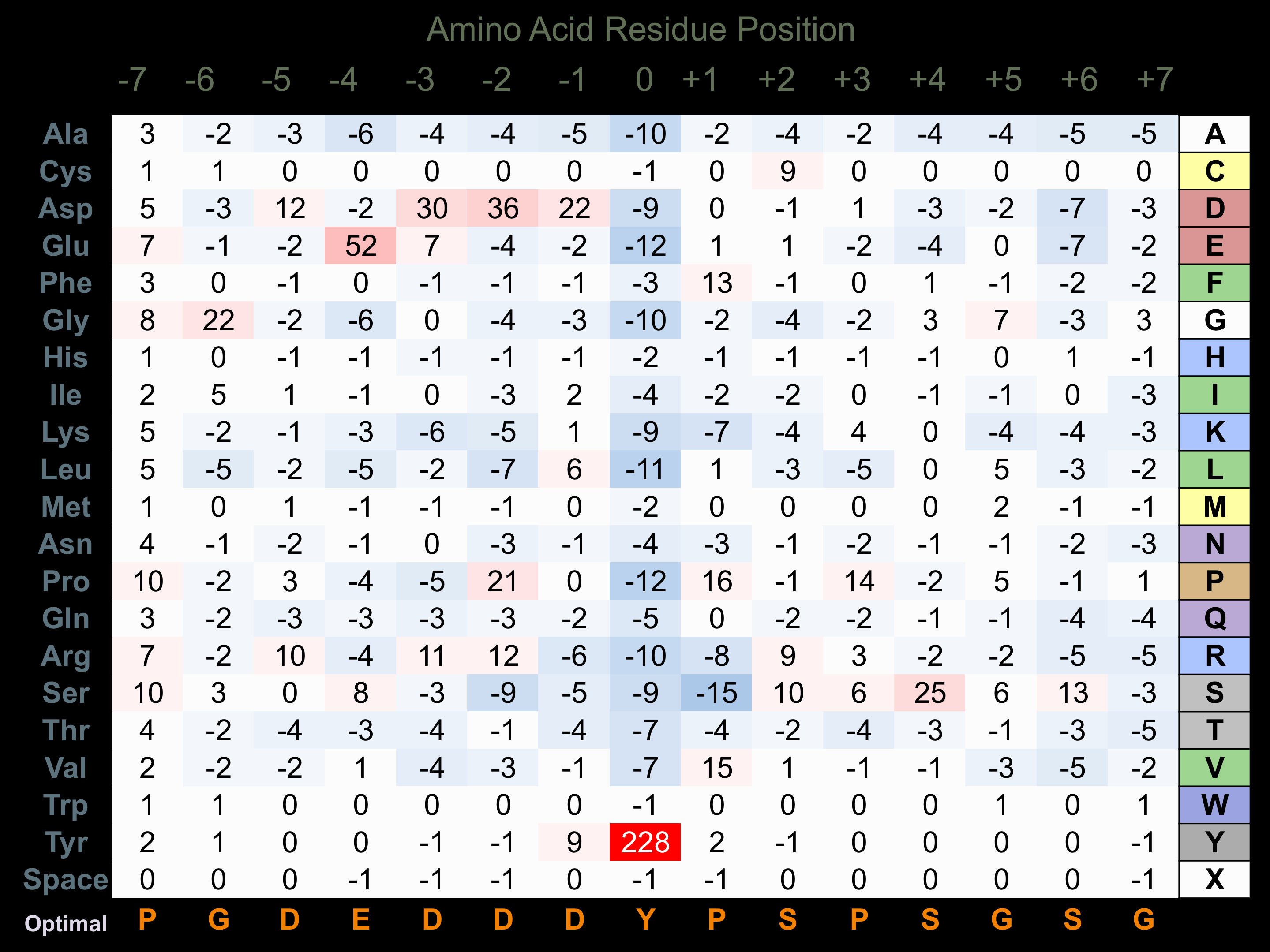
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Prostate cancer
Comments:
In a large 2-stage genomewide association study of prostate cancer, SNP in intron of the LMR gene is associated susceptibility to prostate cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +378, p<0.041); Pituitary adenomas (aldosterone-secreting) (%CFC= -64, p<0.047); and Prostate cancer (%CFC= -49, p<0.024). The COSMIC website notes an up-regulated expression score for LMR2 in diverse human cancers of 623, which is 1.4-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 26 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25425 diverse cancer specimens. This rate is only -8 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.31 % in 895 skin cancers tested; 0.28 % in 589 stomach cancers tested; 0.28 % in 1270 large intestine cancers tested; 0.23 % in 603 endometrium cancers tested; 0.11 % in 1957 lung cancers tested; 0.09 % in 1276 kidney cancers tested; 0.07 % in 757 oesophagus cancers tested; 0.07 % in 548 urinary tract cancers tested; 0.07 % in 273 cervix cancers tested; 0.07 % in 1512 liver cancers tested; 0.06 % in 238 bone cancers tested; 0.05 % in 127 biliary tract cancers tested; 0.04 % in 939 prostate cancers tested; 0.04 % in 1490 breast cancers tested; 0.04 % in 1467 pancreas cancers tested; 0.03 % in 382 soft tissue cancers tested; 0.01 % in 942 upper aerodigestive tract cancers tested; 0.01 % in 891 ovary cancers tested; 0.01 % in 2082 central nervous system cancers tested; 0 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
None > 3 in 19,690 cancer specimens
Comments:
Only 4 deletions, 1 insertion and no complex mutations are noted on the COSMIC website.

