Nomenclature
Short Name:
CDK5
Full Name:
Cyclin-dependent kinase 5
Alias:
- Cell division protein kinase 5
- CRK6
- EC 2.7.11.22
- PSSALRE
- TAU protein kinase II
- TPKII
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDK5
Specific Links
Structure
Mol. Mass (Da):
33,304
# Amino Acids:
292
# mRNA Isoforms:
2
mRNA Isoforms:
33,304 Da (292 AA; Q00535); 29,544 Da (260 AA; Q00535-2)
4D Structure:
Heterodimer of a catalytic subunit and a regulatory subunit (p35). Found in a trimolecular complex with CABLES1 and ABL1. Interacts with CABLES1 By similarity. Interacts with AATK.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 4 | 286 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K9, K33, K56.
Serine phosphorylated:
S46, S72, S105, S159+.
Threonine phosphorylated:
T14-, T17, T231.
Tyrosine phosphorylated:
Y15-, Y158+, Y167-, Y239, Y242.
Ubiquitinated:
K128, K141, K237.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 50
50
812
19
1295
 3
3
51
12
56
 2
2
32
1
0
 35
35
570
54
549
 47
47
757
12
659
 6
6
91
43
122
 31
31
494
23
648
 100
100
1612
28
2602
 19
19
304
10
310
 7
7
114
58
95
 2
2
29
17
26
 30
30
486
127
607
 1
1
16
12
8
 0.6
0.6
9
7
3
 2
2
28
11
30
 6
6
90
9
26
 12
12
199
184
2339
 1
1
16
6
6
 1.1
1.1
17
49
20
 28
28
451
56
489
 5
5
80
14
61
 3
3
41
16
33
 10
10
158
10
83
 1
1
16
8
11
 3
3
49
11
28
 50
50
811
31
1485
 2
2
36
15
20
 0.7
0.7
12
8
6
 1.2
1.2
19
8
13
 4
4
60
14
47
 58
58
927
18
674
 28
28
448
21
526
 63
63
1023
56
1122
 49
49
785
31
690
 2
2
34
22
39
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 59.1
59.1
75.2
100 58.8
58.8
75.2
100 -
-
-
100 -
-
-
96 86.7
86.7
87.9
100 -
-
-
- 99.7
99.7
100
100 99.3
99.3
99.7
99 -
-
-
- -
-
-
- 97.3
97.3
99
97 96.9
96.9
99
97 96.6
96.6
99
97 -
-
-
- 77.5
77.5
86.7
79 81.9
81.9
89.3
- 73.6
73.6
87
75 67.1
67.1
79
- -
-
-
- 57.8
57.8
76.9
- -
-
-
- 56.5
56.5
75.5
- -
-
-
59 -
-
-
62
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
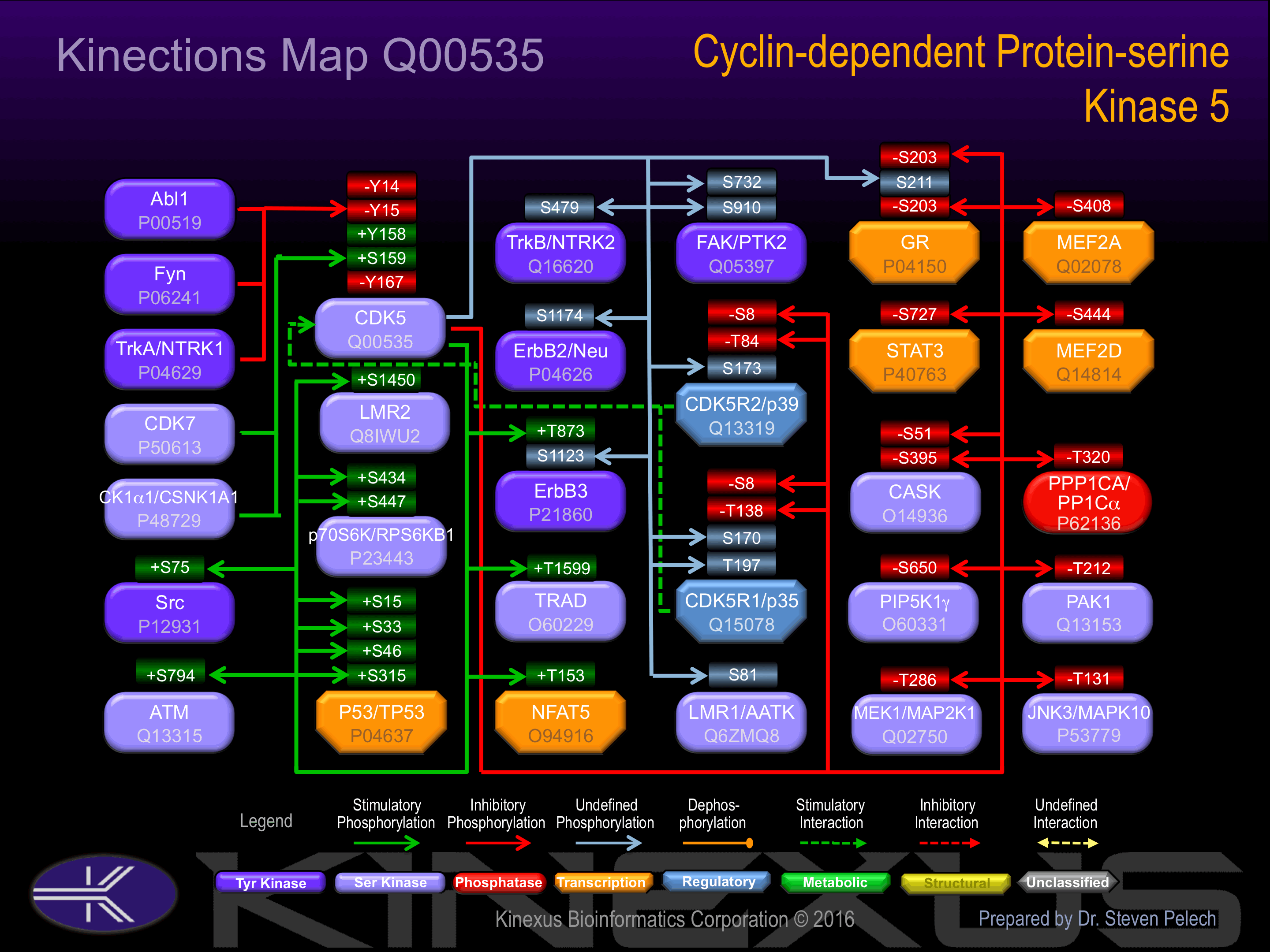
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDK5R1 - Q15078 |
| 2 | MAPT - P10636 |
| 3 | NES - P48681 |
| 4 | NDEL1 - Q9GZM8 |
| 5 | CDK16 - Q00536 |
| 6 | DNM1 - Q05193 |
| 7 | PCNA - P12004 |
| 8 | PURA - Q00577 |
| 9 | PPP1R2P3 - Q6NXS1 |
| 10 | C1orf61 - Q13536 |
| 11 | SET - Q01105 |
| 12 | MAPK10 - P53779 |
| 13 | PAK1 - Q13153 |
| 14 | STX1A - Q16623 |
| 15 | MEF2A - Q02078 |
Regulation
Activation:
Activated by binding p35 protein. Phosphorylation at Ser-159 increases phosphotransferase activity.
Inhibition:
Phosphorylation at Thr-14 and Tyr-15 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Amphiphysin | P49418 | S272 | EEPSPLPSPTASPNH | |
| Amphiphysin | P49418 | S276 | PLPSPTASPNHTLAP | |
| Amphiphysin | P49418 | S285 | NHTLAPASPAPARPR | |
| ATM | Q13315 | S794 | LSNCTKKSPNKIASG | + |
| Bcl-2 | P10415 | S70 | RDPVARTSPLQTPAA | |
| CACNA1C | Q13936 | S783 | KKLARTASPEKKQEL | |
| CASK | O14936 | S395 | DKINTKSSPQIRNPP | - |
| CASK | O14936 | S51 | DVAKFTSSPGLSTED | - |
| CDK5R1 (p35 CDK5 activator) | Q15078 | S170 | CYRLKHLSPTDPVLW | |
| CDK5R1 (p35 CDK5 activator) | Q15078 | S8 | MGTVLSLSPSYRKAT | - |
| CDK5R1 (p35 CDK5 activator) | Q15078 | T138 | PAVISAGTPKRVIVQ | - |
| CDK5R1 (p35 CDK5 activator) | Q15078 | T197 | WQDQGFITPANVVFL | |
| CDK5R2 (p39 CDK5 activator) | Q13319 | S173 | APPVPGGSPRRVIVQ | |
| CDK5R2 (p39 CDK5 activator) | Q13319 | S8 | MGTVLSLSPASSAKG | - |
| CDK5R2 (p39 CDK5 activator) | Q13319 | T84 | KKGSKKVTPKPASTG | - |
| CENTG1 iso2 | Q99490 | S249 | ACKSLPSSPSHSAAS | |
| CENTG1 iso2 | Q99490 | S279 | YSSSLPSSPNVGHRE | |
| CRMP1 | Q14194 | S522 | PAPSAKSSPSKHQPP | |
| CRMP1 | Q14194 | T509 | PVYEVPATPKYATPA | |
| CRMP2 (DPYSL2) | Q16555 | S522 | PASSAKTSPAKQQAP | |
| CRMP4 | Q6DEN2 | S636 | PAGSARGSPTRPNPP | |
| CTNNB1 | P35222 | S191 | SRHAIMRSPQMVSAI | |
| CTNNB1 | P35222 | S246 | ALVKMLGSPVDSVLF | |
| DAB1 | O75553 | S433 | TSDSTRSSPQTDKPR | |
| DAB1 | O75553 | S514 | STTPSTNSPPTPAPR | |
| DAB1 | O75553 | S524 | TPAPRQSSPSKSSAS | |
| DAB1 | O75553 | S548 | IFEEGFESPSKSEEQ | |
| DAB1 | O75553 | S583 | EPSGDNISPQAGS__ | |
| DAB1 | O75553 | T418 | AMFQGPLTPLATVPG | |
| DAB1 | O75553 | T503 | DISQLNLTPVTSTTP | |
| DAB1 | O75553 | T509 | LTPVTSTTPSTNSPP | |
| DAB1 | O75553 | T517 | PSTNSPPTPAPRQSS | |
| DARPP-32 | Q9UD71 | T75 | RPNPCAYTPPSLKAV | |
| DOR1 | P41143 | T161 | VKALDFRTPAKAKLI | |
| Doublecortin | O43602 | S109 | SRMNGLPSPTHSAHC | |
| Doublecortin | O43602 | S287 | AAAGPKASPTPQKTS | |
| Doublecortin | O43602 | S339 | PQKTSAKSPGPMRRS | |
| Doublecortin | O43602 | S368 | ATAGPKASPTPQKTS | |
| Doublecortin | O43602 | S408 | STPKSKQSPISTPTS | |
| Doublecortin | O43602 | S415 | SPISTPTSPGSLRKH | |
| Doublecortin | O43602 | T370 | AGPKASPTPQKTSAK | |
| Doublecortin | O43602 | T402 | TSSSQLSTPKSKQSP | |
| DYN1 | Q05193 | S774 | SVPAGRRSPTSSPTP | |
| DYN1 | Q05193 | S778 | GRRSPTSSPTPQRRA | |
| DYN1 | Q05193 | T780 | RSPTSSPTPQRRAPA | |
| ErbB2 (HER2) | P04626 | S1174 | LERPKTLSPGKNGVV | |
| ErbB3 | P21860 | S1123 | RSRSRSRSPRPRGDS | |
| ErbB3 | P21860 | T873 | LLYSEAKTPIKWMAL | + |
| Ezrin | P15311 | T234 | YEKDDKLTPKIGFPW | |
| FAK (PTK2) | Q05397 | S732 | SSEGFYPSPQHMVQT | ? |
| FAK (PTK2) | Q05397 | S910 | KLQPQEISPPPTANL | ? |
| FZR1 | Q9UM11 | S163 | KSQKLLRSPRKPTRK | |
| FZR1 | Q9UM11 | S40 | PASSPVSSPSKHGDR | |
| FZR1 | Q9UM11 | T121 | DRRLQPSTPEKKGLF | |
| GR | P04150 | S203 | DLEFSSGSPGKETNE | - |
| GR | P04150 | S211 | PGKETNESPWRSDLL | ? |
| GR | P04150 | S226 | IDENCLLSPLAGEDD | - |
| Huntingtin | P42858 | S1181 | LTNPPSLSPIRRKGK | |
| Huntingtin | P42858 | S1201 | EQASVPLSPKKGSEA | |
| JNK3 (MAPK10) | P53779 | T131 | ISLLNVFTPQKTLEE | - |
| LMR1 (AATK) | Q6ZMQ8 | S81 | AADLAQGSPATAAQN | ? |
| LMR2 | Q8IWU2 | S1450 | LQTSKYFSPPPPARS | + |
| MAP1B | P46821 | S2256 | GTKTKSSSPVKKSDG | |
| MAP1B | P46821 | S2289 | DKVSRVASPKKKESV | |
| MAP1B | P46821 | T667 | EVAKKEDTPIKKEEK | |
| MAP2 | P11137 | S136 | ETANLPPSPPPSPAS | |
| MEF2A | Q02078 | S408 | SIKSEPISPPRDRMT | - |
| MEF2D | Q14814 | S444 | SIKSEPVSPSRERSP | - |
| MEK1 (MAP2K1) | Q02750 | T286 | VEGDAAETPPRPRTP | - |
| MUNC18 (STXBP1) | P61764 | T574 | IGSTHILTPQKLLDT | |
| Myc | P01106 | S62 | LLPTPPLSPSRRSGL | |
| NDEL1 | Q9GZM8 | S198 | TRKSAPSSPTLDCEK | |
| NDEL1 | Q9GZM8 | S231 | GTENTFPSPKAIPNG | |
| NDEL1 | Q9GZM8 | T219 | ASLSLPATPVGKGTE | |
| Nestin | P48681 | T315 | AENSRLQTPGGGSKT | |
| nestin iso2 | P48681 | T1299 | GETLPDSTPLGFYLR | |
| NFAT5 | O94916 | T153 | TVQQHPSTPKRHTVL | + |
| NFL (Neurofilament L) | P07196 | T21 | YKRRYVETPRVHISS | |
| NKEF-B | P32119 | T88 | THLAWINTPRKEGGL | |
| NMDAR2A | Q12879 | S1232 | SGHFTMRSPFKCDAC | |
| ODFP1 | Q14990 | S207 | SPCYPCTSPCSPCSP | |
| p53 | P04637 | S15 | PSVEPPLSQETFSDL | + |
| p53 | P04637 | S315 | LPNNTSSSPQPKKKP | + |
| p53 | P04637 | S33 | LPENNVLSPLPSQAM | + |
| p53 | P04637 | S46 | AMDDLMLSPDDIEQW | + |
| p70S6K (RPS6KB1) | P23443 | S434 | SFEPKIRSPRRFIGS | + |
| p70S6K (RPS6KB1) | P23443 | S447 | GSPRTPVSPVKFSPG | + |
| PAK1 | Q13153 | T212 | VIEPLPVTPTRDVAT | - |
| Parkin | O60260 | S131 | HTDSRKDSPPAGSPA | |
| PCTAIRE1 (CDK16, PCTK1) | Q00536 | S95 | TSSDEVQSPVRVRMR | |
| PIP5KG | O60331 | S650 | DERSWVYSPLHYSAQ | - |
| PLD2 | O14939 | S134 | ARFAVAYSPARDAGN | |
| PPP1CA | P62138 | T320 | NPGGRPITPPRNSAK | - |
| PPP1R1A | Q13522 | S6 | __MEPDNSPRKIQFT | |
| PPP1R1A | Q13522 | S67 | LKSTLAMSPRQRKKM | |
| PPP1R2 | P41236 | T72 | MKIDEPSTPYHSMMG | |
| PSEN1 | P49768 | T354 | HLGPHRSTPESRAAV | |
| PXN iso2 | P49023 | S85 | HQQPQSSSPVYGSSA | |
| RasGRF1 | Q13972 | S750 | LSITKTSSPSRRRKL | |
| RasGRF2 | O14827 | S738 | LAVSRTSSPVRARKL | |
| Rb | P06400 | S795 | SPYKFPSSPLRIPGG | - |
| RHOQ | P17081 | T189 | ILTPKKHTVKKRIGS | |
| SDS3 | Q9H7L9 | S228 | RTLNKLKSPKRPASP | |
| Septin-5 | Q99719 | S161 | HCCLYFISPFGHGLR | |
| Septin-5 | Q99719 | S327 | TQDSRMESPIPILPL | |
| Septin-5 iso2 | Q99719-2 | S17 | RLVEQLLSPRTQAQR | |
| SIPA1L1 | O43166 | S1349 | SLGAATSSPRSGPGK | |
| Spinophilin | Q96SB3 | S17 | GGPLRSASPHRSAYE | |
| Src | P12931 | S75 | NSSDTVTSPQRAGPL | + |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| STMN1 | P16949 | S25 | QAFELILSPRSKESV | |
| STMN1 | P16949 | S38 | SVPEFPLSPPKKKDL | |
| SYN1 | P17600 | S553 | ARPPASPSPQRQAGP | |
| SYNJ1 | O43426 | S1163 | REMEAPKSPGTTRKD | |
| Tau | P10636 | S515 | GDRSGYSSPGSPGTP | |
| Tau | P10636 | S518 | SGYSSPGSPGTPGSR | |
| Tau | P10636 | S551 | VVRTPPKSPSSAKSR | |
| Tau | P10636 | S578 | NVKSKIGSTENLKHQ | |
| Tau | P10636 | S712 | GAEIVYKSPVVSGDT | |
| Tau | P10636 | S738 | GSIDMVDSPQLATLA | |
| Tau | P10636 | T497 | KTPPAPKTPPSSGEP | |
| Tau | P10636 | T521 | SSPGSPGTPGSRSRT | |
| Tau | P10636 | T528 | TPGSRSRTPSLPTPP | |
| Tau | P10636 | T533 | SRTPSLPTPPTREPK | |
| Tau | P10636 | T547 | KKVAVVRTPPKSPSS | |
| Tau iso8 | P10636-8 | S409 | DTSPRHLSNVSSTGS | |
| Tau iso9 (Tau-F) | P10636-9 | S404 | PVVSGDTSPRHLSNV | |
| TH | P07101 | S61 | SYTPTPRSPRFIGRR | |
| TPPP | O94811 | S160 | GVTKAISSPTVSRLT | |
| TPPP | O94811 | S18 | ANRTPPKSPGDPSKD | |
| TPPP | O94811 | T14 | PAKAANRTPPKSPGD | |
| TRAD | O60229 | T1599 | EPLQLPKTPAKQRNN | + |
| TrkB (NTRK2) | Q16620 | S479 | SNDDDSASPLHHISN | |
| Vimentin | P08670 | S56 | SRSLYASSPGGVYAT | |
| VR1 | Q8NER1 | T407 | IAYSSSETPNRHDML | |
| WASF2 | Q9Y6W5 | T137 | PPPLNNLTPYRDDGK |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
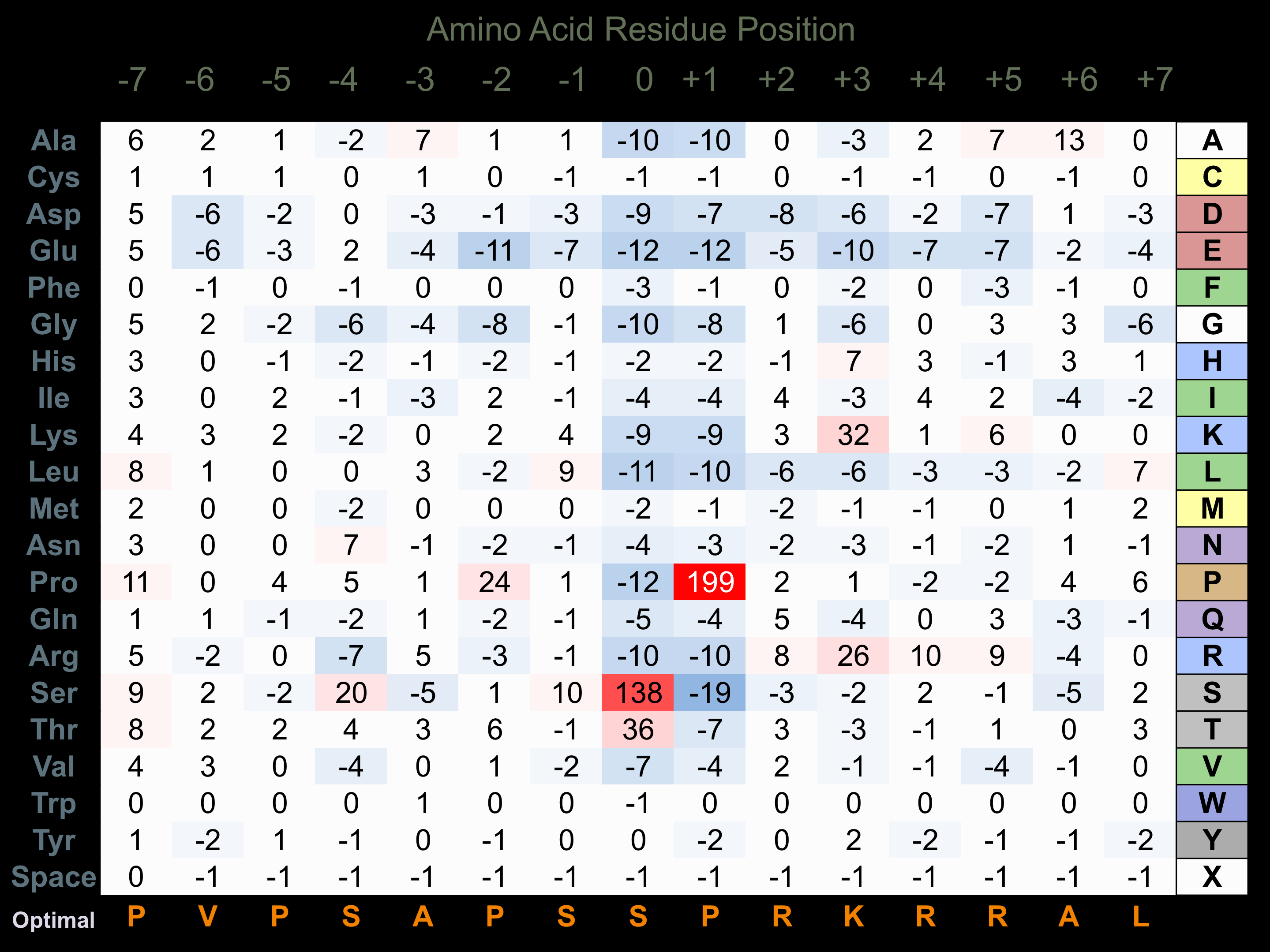
Matrix Type:
Experimentally derived from alignment of 184 known protein substrate phosphosites and 7 peptides phosphorylated by recombinant CDK5 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological, endocrine, and musculoskeletal disorders
Specific Diseases (Non-cancerous):
Alzheimer's disease (AD); Amyotrophic lateral sclerosis (ALS); Neuronitis; Parkinson disease; Diabetes
Comments:
Alzheimer's disease is characterized by an accumulation of p25. CDK5 kinase levels rise with increased p25 levels, but it was not shown to be causative. Binding of p35 or p25 to CDK5 can be impaired with a CDK5 S159T mutation. ALS is a rare motor neuron disease resulting in loss of control over muscle movement. Neuronitis results in loss of motor control, and is associated with BET1 (a golgi vesicular membrane trafficking protein). CDK5 may also contribute to pathogenicity in Parkinson Disease. CDK5 signalling can lead to phosphorylation of a nuclear receptor, PPARG. This CDK5-PPARG pathway has been suggested to have a role in insulin resistance for diabetes.
Specific Cancer Types:
Neuroblastomas (NB); Aneurysmal bone cysts (ABC)
Comments:
CDK5 has been implicated in Aneurysmal bone cyst, which is a bone cancer that will affect bone, endothelial tissue, and bone marrow.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +89, p<0.0001); Cervical cancer (%CFC= -63, p<0.013); Cervical cancer stage 2A (%CFC= +60, p<0.081); Colon mucosal cell adenomas (%CFC= +69, p<0.0001); Large B-cell lymphomas (%CFC= +48, p<0.002); Lung adenocarcinomas (%CFC= +95, p<0.0001); and Oral squamous cell carcinomas (OSCC) (%CFC= +115, p<0.027). The COSMIC website notes an up-regulated expression score for CDK5 in diverse human cancers of 621, which is 1.3-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 28 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24196 diverse cancer specimens. This rate is only -27 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.31 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,222 cancer specimens
Comments:
Only 1 deletion and 1 insertion and no complex mutations are noted on the COSMIC website.