Nomenclature
Short Name:
BRSK1
Full Name:
BR serine/threonine-protein kinase 1
Alias:
- BR serine/threonine kinase 1
- EC 2.7.11.1
- KIAA1811
- SAD1
- SAD1 kinase
- SAD1A
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
BRSK
Structure
Mol. Mass (Da):
86,753
# Amino Acids:
794
# mRNA Isoforms:
3
mRNA Isoforms:
86,753 Da (794 AA; Q8TDC3-2); 85,087 Da (778 AA; Q8TDC3); 38,782 Da (343 AA; Q8TDC3-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
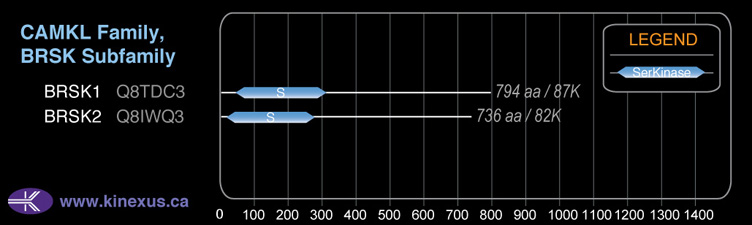
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S193-, S275, S399, S404, S413, S427, S432, S443, S447, S508, S515, S563, S587.
Threonine phosphorylated:
T189+, T417, T583, T772.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1782
9
1707
 19
19
338
6
513
 -
-
-
-
-
 77
77
1381
30
2893
 37
37
658
10
450
 0.8
0.8
14
9
10
 5
5
96
13
56
 45
45
809
7
893
 2
2
30
3
5
 47
47
837
7
891
 27
27
474
7
716
 46
46
819
13
594
 25
25
437
2
356
 18
18
317
5
490
 34
34
598
7
728
 8
8
138
7
175
 17
17
304
11
532
 24
24
427
5
552
 55
55
982
5
1328
 27
27
478
28
217
 83
83
1483
7
2236
 19
19
345
5
391
 -
-
-
-
-
 43
43
772
7
985
 27
27
488
7
458
 36
36
636
21
610
 5
5
81
5
50
 22
22
389
5
512
 6
6
114
5
169
 -
-
-
-
-
 59
59
1043
12
59
 38
38
680
13
2173
 0.2
0.2
4
12
0
 45
45
793
26
415
 14
14
244
22
153
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
99 -
-
-
- -
-
-
99 -
-
-
- 98.1
98.1
98.2
98 -
-
-
- 94.8
94.8
95.4
99 29.2
29.2
44.5
99 -
-
-
- 32.1
32.1
33.3
- 28.3
28.3
43.9
- -
-
-
84 25.1
25.1
38.5
81 -
-
-
- 48.5
48.5
62.5
53 -
-
-
- 25.7
25.7
37.9
34 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 27
27
41.9
- 22.2
22.2
34.2
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC25C - P30307 |
| 2 | CDC25B - P30305 |
| 3 | WEE1 - P30291 |
| 4 | NUAK1 - O60285 |
Regulation
Activation:
Activated by phosphorylation on Thr-205 by STK11 in complex with STE20-related adapter-alpha (STRAD alpha) pseudo kinase and CAB39
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AMPKa1 (PRKAA1) | Q13131 | T183 | SDGEFLRTSCGSPNY | + |
| BRSK1 | Q8TDC3 | S193 | LLETSCGSPHYACPE | - |
| BRSK1 | Q8TDC3 | T189 | VGDSLLETSCGSPHY | + |
| Cdc25B | P30305 | S375 | ARVLRSKSLCHDEIE | |
| Cdc25C | P30307 | S216 | SGLYRSPSMPENLNR | ? |
| MAPT/TAU | P10636 | S579 | NVKSKIGSTENLKHQ | |
| MAPT/TAU | P10636 | T529 | TPGSRSRTPSLPTPP | |
| TUBG1 | P23258 | S131 | READGSDSLEGFVLC | + |
| TUBG2 | Q9NRH3 | S131 | READGSDSLEGFVLC | + |
| Wee1 | P30291 | S642 | KKMNRSVSLTIY___ | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
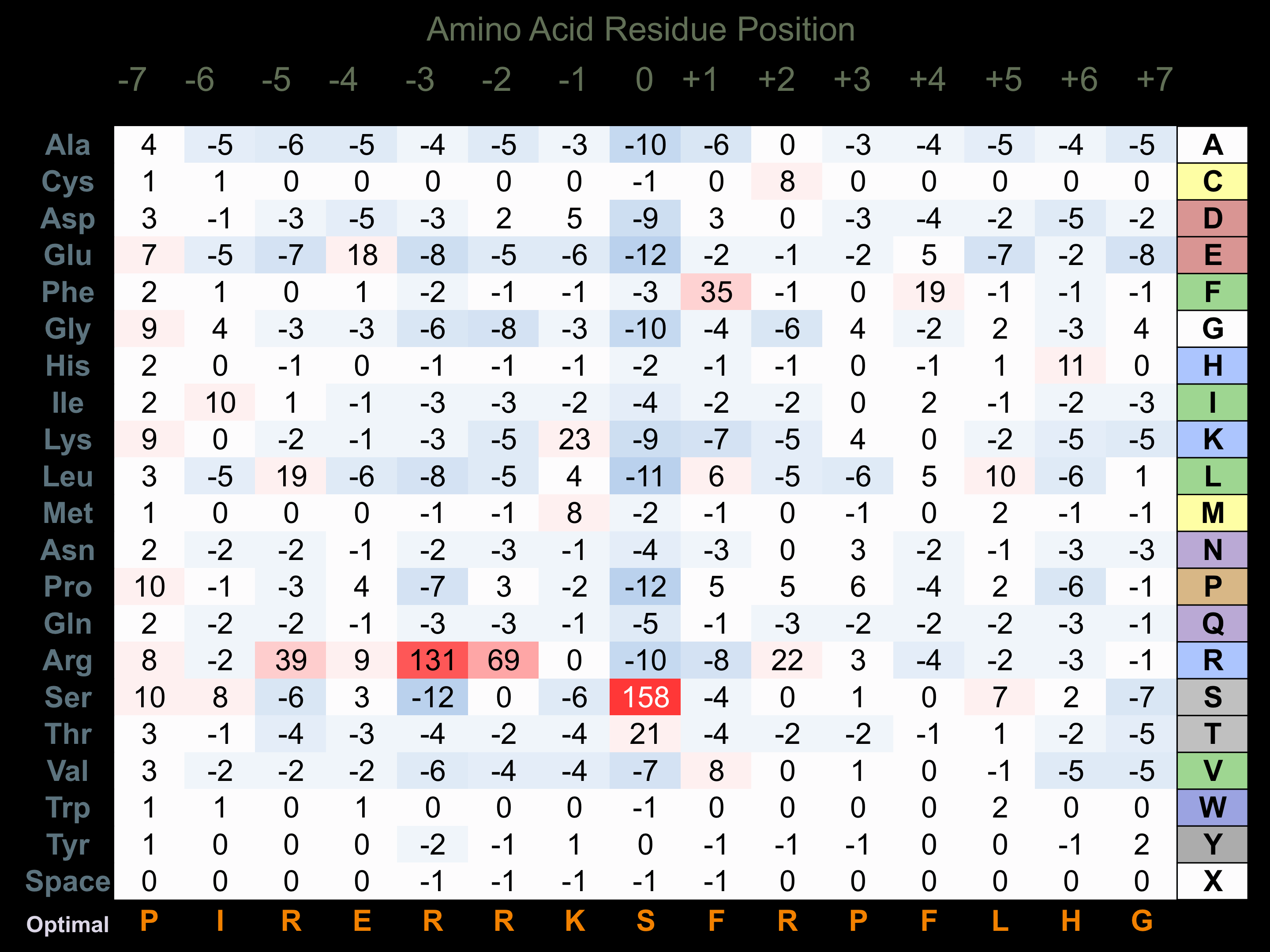
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Menopause
Specific Diseases (Non-cancerous):
Premature ovarian failure (POF); age at natural menopause (AANM)
Comments:
The rs12611091 SNP in BRSK1 can lead to premature ovarian failure, which is a rare condition.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. BRSK1's phosphotransferase activity was lost with the K59A, and G327A mutations.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 24433 diverse cancer specimens. This rate is only 35 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.33 % in 1229 large intestine cancers tested; 0.21 % in 710 oesophagus cancers tested; 0.18 % in 555 stomach cancers tested; 0.16 % in 864 skin cancers tested; 0.14 % in 548 urinary tract cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R303W (5).
Comments:
Twelve R379fs*9 deletions, 9 other deletions, 9 R379fs*54 insertions mutations, and no complex mutations.

