Nomenclature
Short Name:
Wnk4
Full Name:
Serine-threonine-protein kinase WNK4
Alias:
- EC 2.7.11.1
- Kinase WNK4
- PRKWNK4
- Protein kinase with no lysine 4
- Protein kinase, lysine-deficient 4
- WNK lysine deficient protein kinase 4
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
Wnk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
134,739
# Amino Acids:
1243
# mRNA Isoforms:
3
mRNA Isoforms:
134,739 Da (1243 AA; Q96J92); 126,577 Da (1165 AA; Q96J92-3); 73,370 Da (663 AA; Q96J92-2)
4D Structure:
Interacts with the C-terminal region of KCNJ1
1D Structure:
Subfamily Alignment
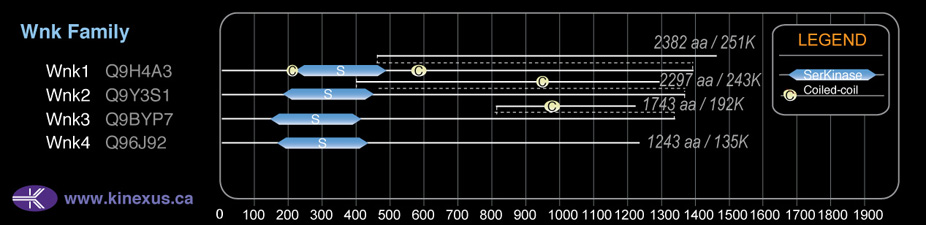
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 174 | 432 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
R124, K226.
Serine phosphorylated:
S47, S64, S97, S219, S239, S331+, S335+, S680, S789, S1190-, S1201, S1217.
Threonine phosphorylated:
T326, T679, T1151, T1154, T1223.
Tyrosine phosphorylated:
Y1113, Y1115, Y1164.
Ubiquitinated:
K328.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 65
65
1521
15
1736
 0.1
0.1
2
5
1
 5
5
110
18
161
 1.2
1.2
28
71
88
 26
26
619
20
442
 0.1
0.1
2
18
2
 2
2
42
23
46
 2
2
51
24
88
 0.1
0.1
3
3
1
 8
8
177
73
224
 4
4
91
23
180
 15
15
362
37
455
 6
6
135
16
179
 0.1
0.1
3
3
0
 0.4
0.4
10
7
9
 0.2
0.2
4
10
4
 2
2
50
108
109
 5
5
108
21
196
 1.2
1.2
27
55
49
 15
15
341
59
223
 3
3
79
25
160
 5
5
108
24
256
 5
5
107
18
172
 7
7
158
19
493
 4
4
100
25
195
 15
15
357
53
417
 8
8
182
18
494
 8
8
180
21
435
 3
3
77
21
120
 6
6
129
28
110
 25
25
584
12
47
 100
100
2342
16
4186
 0.1
0.1
2
24
0
 28
28
667
52
578
 3
3
59
35
55
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 30.6
30.6
38.7
99 30.6
30.6
38.5
97 -
-
-
89 -
-
-
86 77
77
78.5
87 -
-
-
- 86.5
86.5
89.2
88 86.4
86.4
88.9
87 -
-
-
- 27.5
27.5
36.7
- 34.9
34.9
46.5
72 -
-
-
- 46.2
46.2
57.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 21.9
21.9
32.1
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | STK39 - Q9UEW8 |
| 2 | SLC12A3 - P55017 |
| 3 | CLDN4 - O14493 |
| 4 | CLDN1 - O95832 |
| 5 | OXSR1 - O95747 |
| 6 | BCL6 - P41182 |
Regulation
Activation:
Activation requires autophosphorylation of Ser-335. Phosphorylation of Ser-331 also promotes increased activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
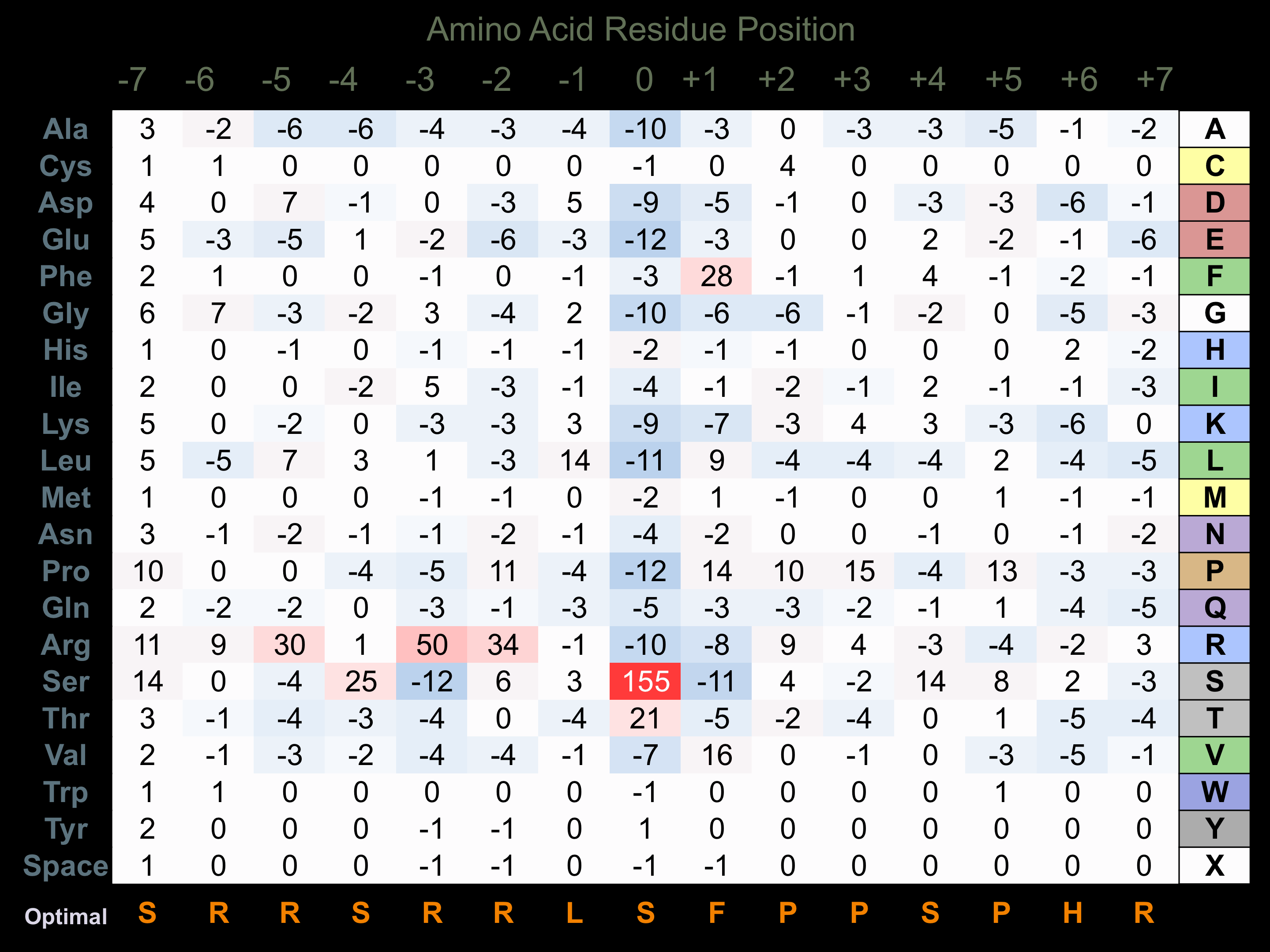
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Nephrological, and metabolic disorders
Specific Diseases (Non-cancerous):
Pseudohypoaldosteronism 2B (PHA2, PHA2B); Pseudohypoaldosteronism (PHA); Essential Hypertension; Pseudohypoaldosteronism Type 1i (DA3); Liddle syndrome; Metabolic acidosis
Comments:
Pseudohypoaldosteronism Type Iib (PHA2, PHA2B) is related to Pseudohypoaldosteronism (PHA) which is a rare nephrological disorder characterized by the lack of response to aldosterone, leading to a lack of feedback inhibition and therefore higher aldosterone levels. PHA can affect the lung, colon, and skin. PHA2B has been characterized by the mutations E562K, D564A, and Q565E each of which can prevent KLHL3 interaction. R1185C is another mutation noted in PHA2B. Essential Hypertension is a rare condition that, through its definition lacks a defined cause. Essential Hypertension is considered to have both a genetic and environmental factor. Pseudohypoaldosteronism Type Ii (DA3) is a rare condition characterized by camptodactylyl, clubfoot, and periodically characterized by cleft palate. DA3 is idiopathic, but is believed to be an x-linked association. Liddle Syndrome is a rare inherited condition characterized by hypertension (high blood pressure). Notably Liddle Syndrome will have lower blood levels of potassium, renin, and aldosterone. Sufferers may have muscle pain, fatigue, weakness, palpitations, or constipation and is caused by mutations in the scnn1b or scnn1g genes.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for WNK4 in diverse human cancers of 399, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24726 diverse cancer specimens. This rate is only -7 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.36 % in 1093 large intestine cancers tested; 0.33 % in 805 skin cancers tested; 0.31 % in 589 stomach cancers tested; 0.16 % in 605 oesophagus cancers tested; 0.15 % in 602 endometrium cancers tested; 0.09 % in 1619 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R669W (4).
Comments:
Twenty-one deletions (17 at V608fs*53), 1 insertion and no complex mutations are noted on the COSMIC website.

