Nomenclature
Short Name:
Wnk1
Full Name:
Serine-threonine-protein kinase WNK1
Alias:
- EC 2.7.11.1
- HSAN2
- MGC163341
- p65
- PRKWNK1
- Protein kinase with no lysine 1; Protein kinase, lysine-deficient 1; PSK
- HSN2
- KDP
- KIAA0344
- MGC163339
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
Wnk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
250794
# Amino Acids:
2382
# mRNA Isoforms:
6
mRNA Isoforms:
299,725 Da (2833 AA; Q9H4A3-7); 279,713 Da (2642 AA; Q9H4A3-6); 279,538 Da (2634 AA; Q9H4A3-5); 250,794 Da (2382 AA; Q9H4A3); 225,560 Da (2135 AA; Q9H4A3-2); 206,646 Da (1975 AA; Q9H4A3-4)
4D Structure:
Interacts with SYT2
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
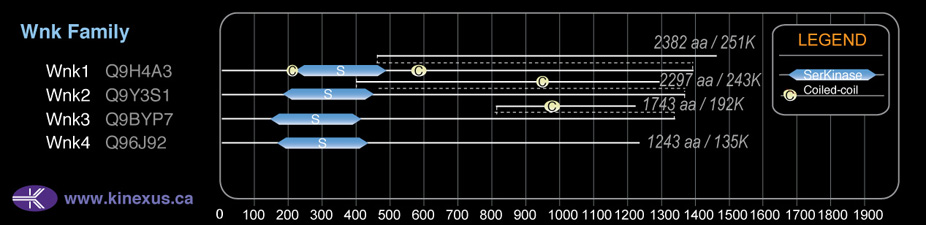
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 194 | 217 | Coiled-coil |
| 221 | 479 | Pkinase |
| 563 | 598 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K1104, K1119, K1128.
Serine phosphorylated:
S11, S15, S19, S33, S167, S170, S174, S183, S185, S189, S198, S378+, S382+, S599, S679, S1105, S1108, S1110, S1116, S1220, S1253, S1261-, S1285, S1295, S1297, S1300, S1849, S1887, S1888, S1890, S1893, S1935, S1959, S1961, S1978, S2002, S2005, S2011, S2012, S2020, S2027, S2029, S2032, S2037, S2038, S2051, S2053, S2121, S2145, S2265, S2270, S2286, S2294, S2370, S2372.
Threonine phosphorylated:
T12, T60+, T74, T373, T1115, T1810, T1848, T1903, T1963, T1968, T1970, T2128, T2245, T2247.
Tyrosine phosphorylated:
Y1215, Y2102, Y2276.
Ubiquitinated:
K375.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 53
53
1007
77
996
 8
8
146
33
163
 42
42
793
23
1143
 32
32
603
267
897
 41
41
778
63
641
 12
12
219
213
434
 9
9
170
81
333
 100
100
1898
112
3447
 33
33
628
38
525
 18
18
344
197
629
 9
9
162
72
262
 42
42
789
381
725
 14
14
264
78
473
 4
4
76
24
91
 4
4
68
38
66
 5
5
95
42
119
 9
9
178
471
1712
 12
12
230
39
380
 18
18
333
203
603
 39
39
745
291
749
 47
47
898
52
1775
 17
17
331
62
648
 18
18
345
36
580
 40
40
764
40
1428
 14
14
273
52
509
 98
98
1869
164
4026
 14
14
271
81
597
 16
16
304
40
503
 21
21
390
40
712
 65
65
1240
70
1365
 29
29
547
42
571
 42
42
791
92
1812
 16
16
309
172
489
 49
49
939
161
751
 12
12
226
87
240
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99
99
99.3
0 96.8
96.8
97.6
97 -
-
-
83 -
-
-
- 89.9
89.9
92.7
91 -
-
-
- 85.5
85.5
89.4
86 77.6
77.6
81.2
86 -
-
-
- 31.4
31.4
44.9
- 69.4
69.4
77.8
73 -
-
-
73.5 37.3
37.3
47.5
60.5 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
41 -
-
-
36 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | SYT2 - Q8N9I0 |
| 2 | AKT1 - P31749 |
| 3 | MAP3K2 - Q9Y2U5 |
| 4 | MAP3K3 - Q99759 |
| 5 | GLIS2 - Q9BZE0 |
| 6 | OXSR1 - O95747 |
| 7 | ME2 - P23368 |
| 8 | SYT1 - P21579 |
| 9 | SYT9 - Q86SS6 |
| 10 | SYT5 - O00445 |
| 11 | SYT6 - Q5T7P8 |
| 12 | SYT10 - Q6XYQ8 |
| 13 | SYT7 - O43581 |
Regulation
Activation:
By hypertonicity. Activation requires autophosphorylation of Ser-382. Phosphorylation of Ser-378 also promotes increased activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| NEDD4L | Q96PU5 | S342 | SSRLRSCSVTDAVAE | - |
| NEDD4L | Q96PU5 | S449 | RRPRSLSSPTVTLSA | |
| OSR1 | O95747 | S325 | VRRVPGSSGRLHKTE | + |
| OSR1 | O95747 | T185 | TRNKVRKTFVGTPCW | + |
| STLK3 (STK39; SPAK) | Q9UEW8 | S373 | VRRVPGSSGHLHKTE | + |
| STLK3 (STK39; SPAK) | Q9UEW8 | T233 | TRNKVRKTFVGTPCW | + |
| SYT2 | Q8N9I0 | S373 | VRRVPGSSGHLHKTE | |
| SYT2 | Q8N9I0 | T199 | ETKVHRKTLNPAFNE | |
| Wnk1 (PRKWNK1) | Q9H4A3 | S378 | LATLKRASFAKSVIG | + |
| Wnk1 (PRKWNK1) | Q9H4A3 | S382 | KRASFAKSVIGTPEF | + |
| Wnk4 (PRKWNK4) | Q96J92 | S335 | KRASFAKSVIGTPEF | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
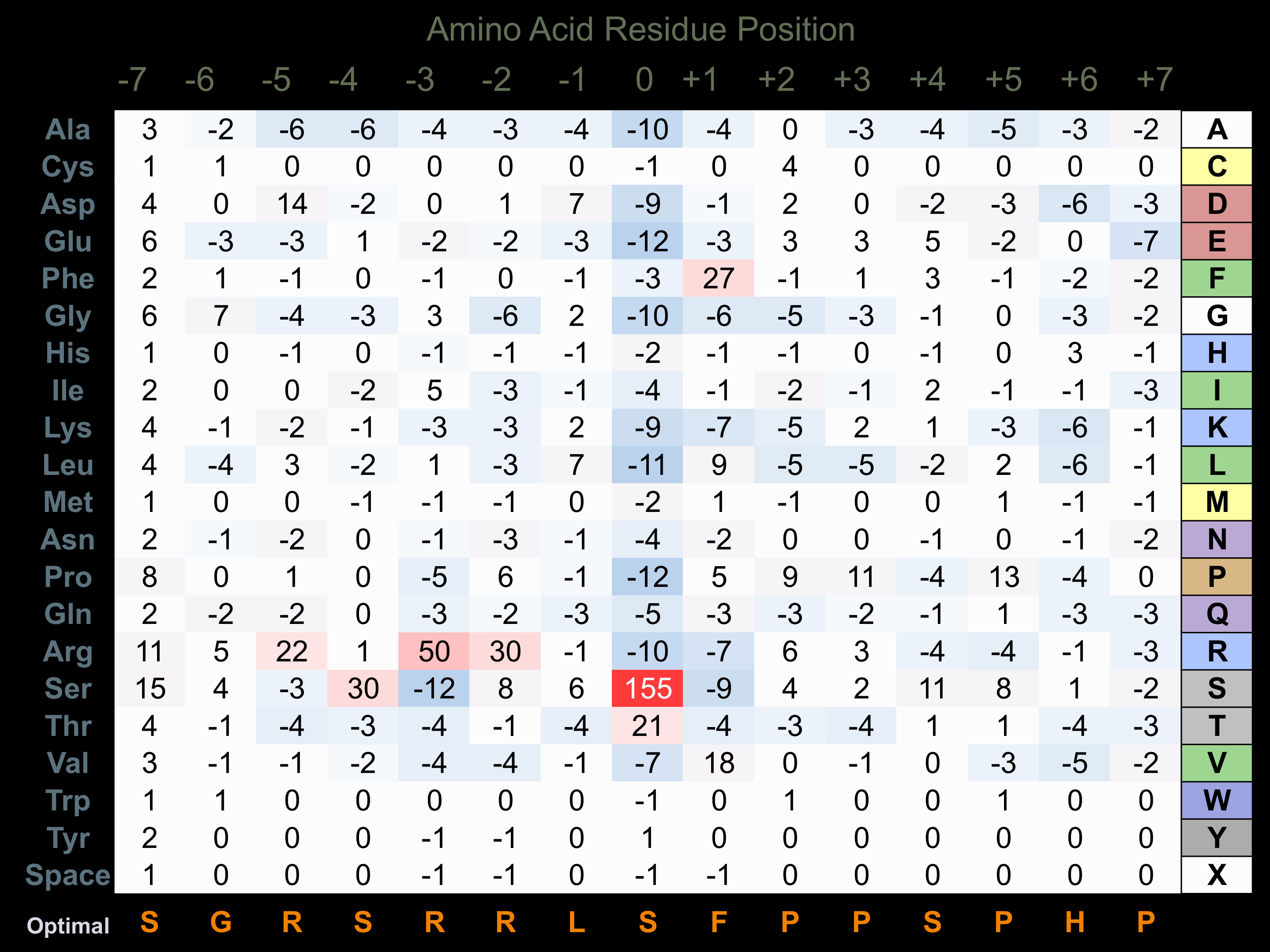
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Neurological, and nephrological disorders
Specific Diseases (Non-cancerous):
Neuropathy; Hereditary sensory neuropathy (CIP, HSAN, HSN); Pseudohypoaldosteronism (PHA) ; Hereditary sensory and autonomic neuropathy Type 1i (HSAN2, HSAN2A); Pseudohypoaldosteronism Type 1i (DA3); Autonomic neuropathy; Pseudohypoaldosteronism Type 1ic; Gitelman syndrome (GS); Hereditary sensory and autonomic neuropathy Type 1ia; Pseudohypoaldosteronism Type 1ib (PHA2)
Comments:
Hereditary Sensory Neuropathy (CIP, HSAN, HSN) is a rare condition that is characterized by congenital insensitivity to pain, and is a disorder which can affect bone, spinal cord, and brain. Pseudohypoaldosteronism (PHA) is a rare nephrological disorder characterized by the lack of response to aldosterone, leading to a lack of feedback inhibition and therefore higher aldosterone levels. PHA can affect the lung, colon, and skin. Hereditary Sensory and Autonomic Neuropathy Type Ii (HSAN2, HSAN2A) is a rare condition that affects sensory neurons, and rarely affects autonomic nervous system nerves leading to abnormal heart rate, digestion, and breathing. HSAN2 can affect heart and bone. Pseudohypoaldosteronism Type Ii (DA3) is a rare condition characterized by camptodactylyl, clubfoot, and periodicially characterized by cleft palate. DA3 is idiopathic, but is believed to be an x-linked association. Pseudohypoaldosteronism Type Iic is a rare condition related to Pseudohypoaldosteronism Type Iib. Gitelman Syndrome (GS) is a rare condition leading to the imbalance of potassium, calcium, and magnesium. Hereditary Sensory and Autonomic Neuropathy Type Iia is a rare condition. Pseudohypoaldosteronism Type Iib (PHA2) is related to Pseudohypoaldosteronism.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -45, p<0.039); Bladder carcinomas (%CFC= +56, p<0.015); Brain glioblastomas (%CFC= +84, p<0.054); Cervical cancer (%CFC= +81, p<0.006); Colorectal adenocarcinomas (early onset) (%CFC= +58, p<0.038); and Gastric cancer (%CFC= -65, p<0.026). The COSMIC website notes an up-regulated expression score for WNK1 in diverse human cancers of 527, which is 1.1-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 62 for this protein kinase in human cancers was 1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25303 diverse cancer specimens. This rate is only -22 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.3 % in 1119 large intestine cancers tested; 0.21 % in 589 stomach cancers tested; 0.2 % in 805 skin cancers tested; 0.15 % in 602 endometrium cancers tested; 0.11 % in 500 urinary tract cancers tested; 0.1 % in 1753 lung cancers tested; 0.05 % in 904 ovary cancers tested; 0.05 % in 1560 breast cancers tested; 0.05 % in 1270 liver cancers tested; 0.04 % in 1226 kidney cancers tested; 0.02 % in 1942 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
None > 7 in 20,587 cancer specimens
Comments:
Twenty-two deletions (19 at K583fs*11), 4 insertions and no complex mutations are noted on the COSMIC website.

