Nomenclature
Short Name:
ChaK1
Full Name:
Transient receptor potential cation channel subfamily M member 7
Alias:
- EC 2.7.11.1
- Long transient receptor potential channel 7
- LTRPC7
- TRPM7
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
Alpha
SubFamily:
ChaK
Specific Links
Structure
Mol. Mass (Da):
212,697
# Amino Acids:
1865
# mRNA Isoforms:
1
mRNA Isoforms:
212,697 Da (1865 AA; Q96QT4)
4D Structure:
Homodimer. Interacts with PLCB1 By similarity. Forms heterodimers with TRPM6.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
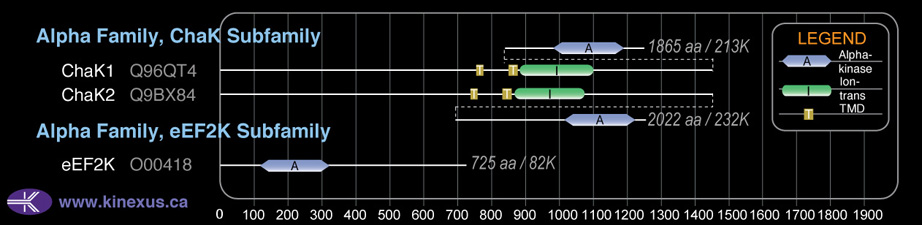
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 757 | 774 | TMD |
| 853 | 875 | TMD |
| 884 | 1096 | Ion_trans |
| 1594 | 1824 | Alpha_kinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
M1, K1168.
Serine phosphorylated:
S57, S101, S764, S907, S1191, S1193, S1230, S1239, S1255, S1258, S1271, S1287, S1358, S1386, S1387, S1390, S1395, S1396, S1404, S1407, S1410, S1413, S1446, S1456, S1463, S1468, S1476, S1477, S1488, S1493, S1500, S1504, S1513, S1527, S1533, S1543, S1567, S1569, S1598, S1615, S1660, S1695, S1779, S1851, S1860.
Threonine phosphorylated:
T10, T523, T555, T1163, T1265, T1405, T1417, T1419, T1435, T1455, T1471, T1482, T1505, T1508, T1537, T1542, T1551, T1583, T1685, T1830.
Tyrosine phosphorylated:
Y104, Y776.
Ubiquitinated:
K667.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 69
69
1839
39
934
 6
6
154
11
318
 -
-
-
-
-
 5
5
130
157
510
 27
27
711
56
473
 0.8
0.8
22
54
17
 1.2
1.2
31
63
37
 65
65
1738
20
3656
 0.2
0.2
6
3
2
 3
3
79
133
122
 5
5
138
13
230
 27
27
717
60
565
 35
35
935
2
146
 21
21
571
5
884
 6
6
158
13
309
 3
3
77
32
254
 2
2
66
230
86
 27
27
712
5
990
 3
3
83
95
311
 27
27
715
168
523
 8
8
201
13
404
 1.4
1.4
38
11
32
 -
-
-
-
-
 35
35
938
7
1361
 4
4
97
13
115
 31
31
840
109
987
 14
14
370
5
569
 22
22
583
5
838
 21
21
573
5
804
 5
5
132
70
243
 31
31
825
12
106
 100
100
2680
38
6892
 9
9
242
105
517
 28
28
739
156
611
 2
2
43
87
36
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.9
100 50.1
50.1
64.1
- -
-
-
96 -
-
-
- 96.5
96.5
98
97 -
-
-
- 94.2
94.2
97.1
94 93.7
93.7
96.8
94 -
-
-
- -
-
-
- 87
87
92.7
88 -
-
-
80 72.2
72.2
83.1
76 -
-
-
- 33.3
33.3
51.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TRPM6 - Q9BX84 |
| 2 | ANXA1 - P04083 |
| 3 | MYH10 - P35580 |
| 4 | MYH14 - Q7Z406 |
| 5 | MYH9 - P35579 |
| 6 | PLCG1 - P19174 |
| 7 | PLCB2 - Q00722 |
| 8 | PLCB1 - Q9NQ66 |
| 9 | PLCB3 - Q01970 |
| 10 | EEF2 - P13639 |
| 11 | MBP - P02686 |
| 12 | PTEN - P60484 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ChaK1 | Q96QT4 | T1163 | DGPKLFLTEEDQKKL | |
| ChaK1 | Q96QT4 | S1191 | EKDDKFHSGSEERIR | |
| ChaK1 | Q96QT4 | S1193 | DDKFHSGSEERIRVT | |
| ChaK1 | Q96QT4 | S1255 | TLTAQKASEASKVHN | |
| ChaK1 | Q96QT4 | S1258 | AQKASEASKVHNEIT | |
| ChaK1 | Q96QT4 | T1265 | SKVHNEITRELSISK | |
| ChaK1 | Q96QT4 | S1271 | ITRELSISKHLAQNL | |
| ChaK1 | Q96QT4 | S1287 | DDGPVRPSVWKKHGV | |
| ChaK1 | Q96QT4 | S1358 | SSGALFPSAVSPPEL | |
| ChaK1 | Q96QT4 | S1387 | KNQKLGSSSTSIPHL | |
| ChaK1 | Q96QT4 | S1390 | KLGSSSTSIPHLSSP | |
| ChaK1 | Q96QT4 | S1404 | PPTKFFVSTPSQPSC | |
| ChaK1 | Q96QT4 | T1405 | PTKFFVSTPSQPSCK | |
| ChaK1 | Q96QT4 | S1407 | KFFVSTPSQPSCKSH | |
| ChaK1 | Q96QT4 | T1435 | KATEGDNTEFGAFVG | |
| ChaK1 | Q96QT4 | S1446 | AFVGHRDSMDLQRFK | |
| ChaK1 | Q96QT4 | T1455 | DLQRFKETSNKIKIL | |
| ChaK1 | Q96QT4 | S1456 | LQRFKETSNKIKILS | |
| ChaK1 | Q96QT4 | S1463 | SNKIKILSNNNTSEN | |
| ChaK1 | Q96QT4 | S1468 | ILSNNNTSENTLKRV | |
| ChaK1 | Q96QT4 | T1471 | NNNTSENTLKRVSSL | |
| ChaK1 | Q96QT4 | S1476 | ENTLKRVSSLAGFTD | |
| ChaK1 | Q96QT4 | S1477 | NTLKRVSSLAGFTDC | |
| ChaK1 | Q96QT4 | T1482 | VSSLAGFTDCHRTSI | |
| ChaK1 | Q96QT4 | S1493 | RTSIPVHSKQAEKIS | |
| ChaK1 | Q96QT4 | T1505 | KISRRPSTEDTHEVD | |
| ChaK1 | Q96QT4 | T1508 | RRPSTEDTHEVDSKA | |
| ChaK1 | Q96QT4 | S1513 | EDTHEVDSKAALIPD | |
| ChaK1 | Q96QT4 | S1527 | DWLQDRPSNREMPSE | |
| ChaK1 | Q96QT4 | S1533 | PSNREMPSEEGTLNG | |
| ChaK1 | Q96QT4 | T1537 | EMPSEEGTLNGLTSP | |
| ChaK1 | Q96QT4 | T1542 | EGTLNGLTSPFKPAM | |
| ChaK1 | Q96QT4 | S1543 | GTLNGLTSPFKPAMD | |
| ChaK1 | Q96QT4 | T1551 | PFKPAMDTNYYYSAV | |
| ChaK1 | Q96QT4 | S1567 | RNNLMRLSQSIPFTP | |
| ChaK1 | Q96QT4 | T1583 | PPRGEPVTVYRLEES | |
| ChaK1 | Q96QT4 | S1598 | SPNILNNSMSSWSQL | |
| ChaK1 | Q96QT4 | S1615 | CAKIEFLSKEEMGGG | |
| ChaK1 | Q96QT4 | S1660 | EVVNTWSSIYKEDTV | |
| ChaK1 | Q96QT4 | T1685 | QRAAQKLTFAFNQMK | |
| ChaK1 | Q96QT4 | S1695 | FNQMKPKSIPYSPRF | |
| ChaK1 | Q96QT4 | S1779 | GENLTDPSVIKAEEK | |
| ChaK1 | Q96QT4 | T1830 | DLKRNDYTPDKIIFP | |
| ChaK1 | Q96QT4 | S1851 | LNLQPGNSTKESEST | |
| ChaK1 | Q96QT4 | S1860 | KESESTNSVRLML__ |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Chak1 | Q96QT4 | S1456 | LQRFKETSNKIKILS | |
| Chak1 | Q96QT4 | S1463 | SNKIKILSNNNTSEN | |
| Chak1 | Q96QT4 | S1468 | ILSNNNTSENTLKRV | |
| Chak1 | Q96QT4 | S1476 | ENTLKRVSSLAGFTD | |
| Chak1 | Q96QT4 | S1477 | NTLKRVSSLAGFTDC | |
| Chak1 | Q96QT4 | S1493 | RTSIPVHSKQAEKIS | |
| Chak1 | Q96QT4 | S1513 | EDTHEVDSKAALIPD | |
| Chak1 | Q96QT4 | S1527 | DWLQDRPSNREMPSE | |
| Chak1 | Q96QT4 | S1533 | PSNREMPSEEGTLNG | |
| Chak1 | Q96QT4 | S1543 | GTLNGLTSPFKPAMD | |
| Chak1 | Q96QT4 | S1567 | RNNLMRLSQSIPFTP | |
| Chak1 | Q96QT4 | S1598 | SPNILNNSMSSWSQL | |
| Chak1 | Q96QT4 | S1615 | CAKIEFLSKEEMGGG | |
| Chak1 | Q96QT4 | S1660 | EVVNTWSSIYKEDTV | |
| Chak1 | Q96QT4 | S1695 | FNQMKPKSIPYSPRF | |
| Chak1 | Q96QT4 | S1779 | GENLTDPSVIKAEEK | |
| Chak1 | Q96QT4 | S1851 | LNLQPGNSTKESEST | |
| Chak1 | Q96QT4 | S1860 | KESESTNSVRLML__ | |
| Chak1 | Q96QT4 | T1163 | DGPKLFLTEEDQKKL | |
| Chak1 | Q96QT4 | T1265 | SKVHNEITRELSISK | |
| Chak1 | Q96QT4 | T1405 | PTKFFVSTPSQPSCK | |
| Chak1 | Q96QT4 | T1435 | KATEGDNTEFGAFVG | |
| Chak1 | Q96QT4 | T1455 | DLQRFKETSNKIKIL | |
| Chak1 | Q96QT4 | T1471 | NNNTSENTLKRVSSL | |
| Chak1 | Q96QT4 | T1482 | VSSLAGFTDCHRTSI | |
| Chak1 | Q96QT4 | T1505 | KISRRPSTEDTHEVD | |
| Chak1 | Q96QT4 | T1508 | RRPSTEDTHEVDSKA | |
| Chak1 | Q96QT4 | T1537 | EMPSEEGTLNGLTSP | |
| Chak1 | Q96QT4 | T1542 | EGTLNGLTSPFKPAM | |
| Chak1 | Q96QT4 | T1551 | PFKPAMDTNYYYSAV | |
| Chak1 | Q96QT4 | T1583 | PPRGEPVTVYRLEES | |
| Chak1 | Q96QT4 | T1685 | QRAAQKLTFAFNQMK | |
| Chak1 | Q96QT4 | T1830 | DLKRNDYTPDKIIFP | |
| MYH9 | P35579 | S1802 | EMEGTVKSKYKASIT | |
| MYH9 | P35579 | S1803 | EMEGTVKSKYKASIT | |
| MYH9 | P35579 | S1808 | VKSKYKASITALEAK | |
| MYH9 | P35579 | T1800 | KLQEMEGTVKSKYKA | |
| MYH9 | P35579 | T1930 | GDLPFVVTRRIVRKG | |
| eEF2K | O00418 | S78 | SSGSPANSFHFKEAW | - |
| ANXA1 | P04083 | S4 | ____AMVSEFLKQAW | |
| Chak1 | Q96QT4 | S1191 | EKDDKFHSGSEERIR | |
| Chak1 | Q96QT4 | S1193 | DDKFHSGSEERIRVT | |
| Chak1 | Q96QT4 | S1255 | TLTAQKASEASKVHN | |
| Chak1 | Q96QT4 | S1258 | AQKASEASKVHNEIT | |
| Chak1 | Q96QT4 | S1271 | ITRELSISKHLAQNL | |
| Chak1 | Q96QT4 | S1287 | DDGPVRPSVWKKHGV | |
| Chak1 | Q96QT4 | S1358 | SSGALFPSAVSPPEL | |
| Chak1 | Q96QT4 | S1387 | KNQKLGSSSTSIPHL | |
| Chak1 | Q96QT4 | S1390 | KLGSSSTSIPHLSSP | |
| Chak1 | Q96QT4 | S1404 | PPTKFFVSTPSQPSC | |
| Chak1 | Q96QT4 | S1407 | KFFVSTPSQPSCKSH | |
| Chak1 | Q96QT4 | S1446 | AFVGHRDSMDLQRFK |
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Amyotrophic lateral sclerosis-Parkinsonism/dementia complex; Amyotrophic lateral sclerosis (ALS); Dementia; lateral sclerosis; Hypomagnesemia with secondary hypocalcemia; Amyotrophic lateral sclerosis-Parkinsonism/dementia Complex 1 (ALS-PDC1)
Comments:
K1648R and G1799D mutations lead to loss of kinase activity and are associated with Amyotrophic lateral sclerosis-parkinsonism/dementia complex 1 (ALS-PDC1), which is characterized by chronic, progressive and uniformly fatal amyotrophic lateral sclerosis and parkinsonism-dementia.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Prostate cancer - metastatic (%CFC= +64, p<0.005). The COSMIC website notes an up-regulated expression score for ChaK1 in diverse human cancers of 308, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 168 for this protein kinase in human cancers was 2.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24783 diverse cancer specimens. This rate is only -33 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.26 % in 1270 large intestine cancers tested; 0.2 % in 864 skin cancers tested; 0.19 % in 589 stomach cancers tested; 0.18 % in 603 endometrium cancers tested; 0.1 % in 710 oesophagus cancers tested; 0.08 % in 1635 lung cancers tested; 0.06 % in 273 cervix cancers tested; 0.05 % in 1512 liver cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: E800K (5); M830V (5); R588Q (4); R274I (4).
Comments:
Only 6 deletions, 2 insertions and no complex mutations are noted on the COSMIC website.

