Nomenclature
Short Name:
PASK
Full Name:
PAS domain containing serine-threonine-protein kinase
Alias:
- EC 2.7.11.1
- KIAA0135
- PASKIN
- PAS-kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
PASK
Specific Links
Structure
Mol. Mass (Da):
142929
# Amino Acids:
1323
# mRNA Isoforms:
3
mRNA Isoforms:
143,684 Da (1330 AA; Q96RG2-2); 142,929 Da (1323 AA; Q96RG2); 123,048 Da (1143 AA; Q96RG2-4)
4D Structure:
NA
1D Structure:
Subfamily Alignment
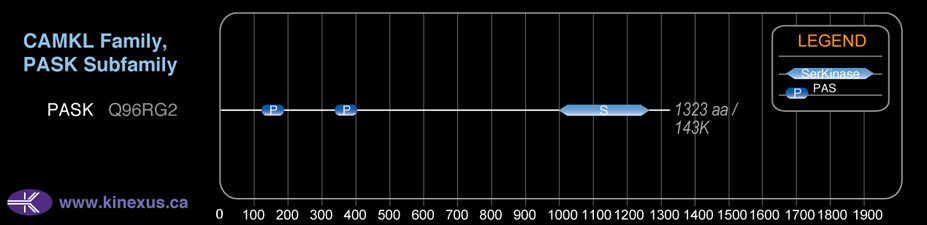
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K218.
Serine phosphorylated:
S19, S40, S55, S60, S104, S106, S109, S116, S390, S524, S533, S579, S582, S667, S670, S939, S949, S1000, S1003, S1057, S1273, S1277, S1280, S1287, S1323.
Threonine phosphorylated:
T34, T642, T874, T954, T1001, T1161+, T1165-, T1322.
Tyrosine phosphorylated:
Y389, Y999.
Ubiquitinated:
K316, K594.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 31
31
837
42
1163
 1.1
1.1
30
17
30
 3
3
72
7
66
 16
16
451
123
905
 19
19
533
33
441
 2
2
66
126
141
 5
5
134
40
327
 47
47
1286
46
2356
 11
11
294
21
163
 2
2
46
105
46
 2
2
57
30
66
 24
24
656
225
614
 4
4
106
41
69
 1.2
1.2
34
11
21
 2
2
65
16
54
 0.9
0.9
26
22
26
 4
4
99
126
387
 2
2
42
16
44
 2
2
42
113
26
 11
11
297
159
336
 2
2
61
20
50
 3
3
80
25
56
 3
3
82
19
62
 5
5
129
16
100
 5
5
145
20
128
 43
43
1188
72
1915
 4
4
114
41
77
 2
2
58
16
61
 2
2
53
16
45
 4
4
119
42
129
 33
33
905
30
843
 100
100
2739
51
6662
 7
7
180
83
367
 32
32
872
83
791
 7
7
194
48
160
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.4
98.4
98.9
98 -
-
-
- -
-
-
- -
-
-
- 65.7
65.7
73.4
70 -
-
-
- 69.7
69.7
77.4
73 67.4
67.4
76.9
71.5 -
-
-
- 46.4
46.4
58.5
- 37.6
37.6
48
64 -
-
-
42 23.8
23.8
39.5
- -
-
-
- 21.4
21.4
37.8
43 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PSMB1 - P20618 |
| 2 | FBL - P22087 |
Regulation
Activation:
Activated by autophosphorylation on Thr-1161 and Thr-1165
Inhibition:
inhibited by the first PAS domain.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
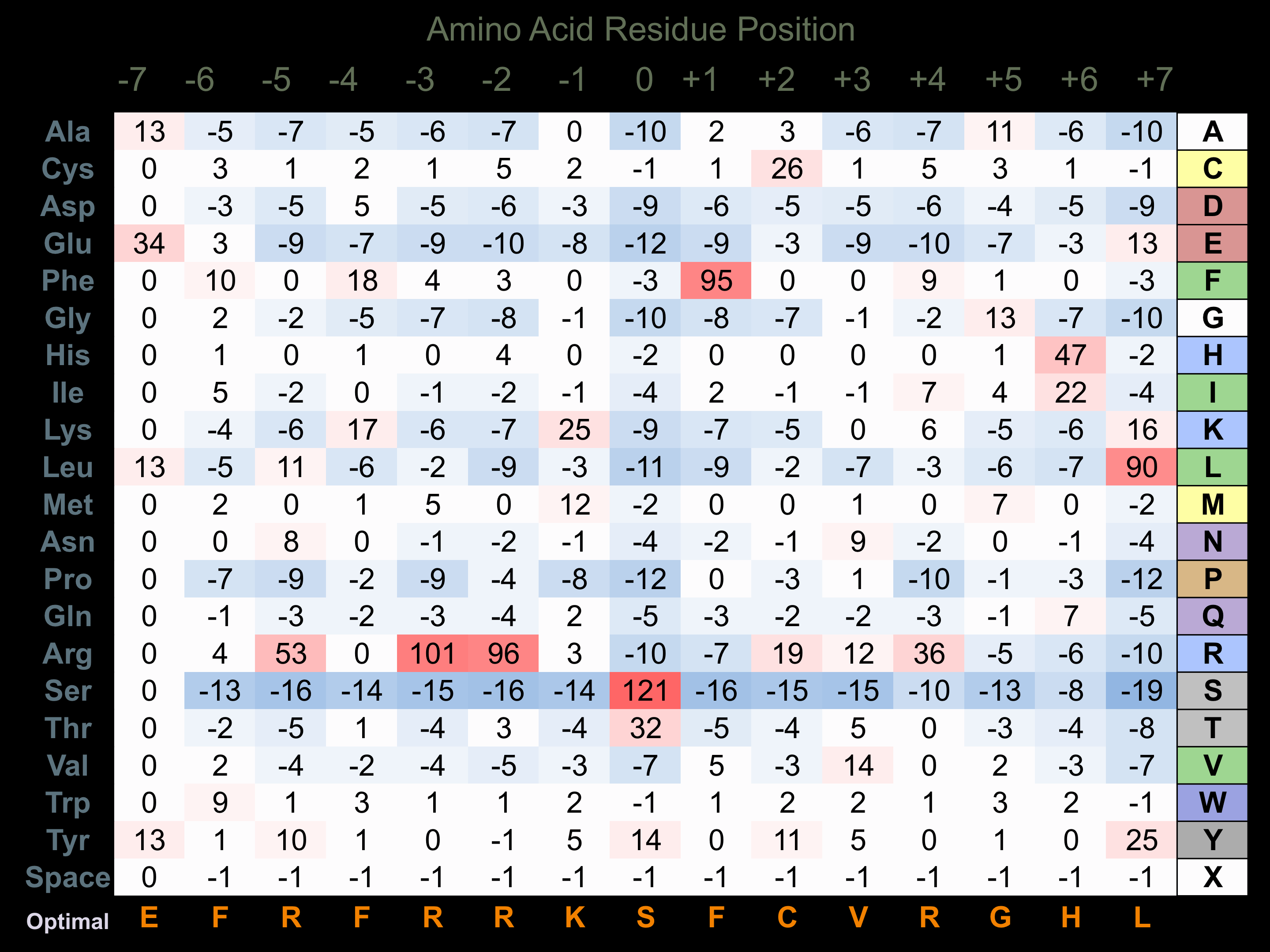
Matrix Type:
Experimentally derived from alignment of 6 known protein substrate phosphosites and 22 peptides phosphorylated by recombinant PASK in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | IC50 < 25 nM | 5279 | 22037377 | |
| AT9283 | IC50 > 100 nM | 24905142 | 19143567 | |
| K-252a; Nocardiopsis sp. | IC50 > 150 nM | 3813 | 281948 | 22037377 |
| TBCA | IC50 > 150 nM | 1095828 | 22037377 | |
| AG-E-60384 | IC50 > 250 nM | 6419741 | 413188 | 22037377 |
| JAK3 Inhibitor VI | IC50 > 250 nM | 16760524 | 22037377 | |
| PKR Inhibitor | IC50 > 250 nM | 6490494 | 235641 | 22037377 |
| SB218078 | IC50 > 250 nM | 447446 | 289422 | 22037377 |
| AS601245 | IC50 > 1 µM | 11422035 | 191384 | 22037377 |
| Bisindolylmaleimide I | IC50 > 1 µM | 2396 | 7463 | 22037377 |
| MK5108 | IC50 > 1 µM | 24748204 | 20053775 | |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| SNS314 | IC50 > 1 µM | 16047143 | 514582 | 18678489 |
| WHI-P154 | IC50 > 1 µM | 3795 | 473773 | 22037377 |
| Momelotinib | IC50 > 2 µM | 25062766 | 19295546 | |
| JNJ-28871063 | IC50 > 4 µM | 17747413 | 17975007 |
Disease Linkage
General Disease Association:
Infectious disease, and diabetes
Specific Diseases (Non-cancerous):
Glanders; Diabetes Type II
Comments:
PASK expression is regulated by glucose, and functions in the regulation of insulin gene expression and glucagon secretion. Downregulation may associate with type 2 diabetes. Mutations are associated with basal insulin hypersecretion.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial carcinomas (%CFC= +129, p<0.011); Classical Hodgkin lymphomas (%CFC= +177, p<0.0001); Large B-cell lymphomas (%CFC= +212, p<(0.0003); and Ovary adenocarcinomas (%CFC= +159, p<0.0009).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24447 diverse cancer specimens. This rate is only -5 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.3 % in 864 skin cancers tested; 0.3 % in 1229 large intestine cancers tested; 0.24 % in 603 endometrium cancers tested; 0.24 % in 589 stomach cancers tested; 0.15 % in 1608 lung cancers tested; 0.08 % in 1289 breast cancers tested; 0.07 % in 710 oesophagus cancers tested; 0.06 % in 807 ovary cancers tested; 0.06 % in 548 urinary tract cancers tested; 0.06 % in 273 cervix cancers tested; 0.06 % in 1253 kidney cancers tested; 0.05 % in 881 prostate cancers tested; 0.04 % in 558 thyroid cancers tested; 0.03 % in 942 upper aerodigestive tract cancers tested; 0.03 % in 238 bone cancers tested; 0.03 % in 1512 liver cancers tested; 0.02 % in 441 autonomic ganglia cancers tested; 0.02 % in 1982 haematopoietic and lymphoid cancers tested; 0.02 % in 1437 pancreas cancers tested; 0.01 % in 2030 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R880H (4); E100K (3).
Comments:
Only 4 deletions, and no insertions or complex mutations are noted on the COSMIC website.

