Nomenclature
Short Name:
PRPK
Full Name:
TP53 regulating kinase
Alias:
- BUD32
- Nori-2
- Nori-2p
- P53-related protein kinase
- Prpk
- TP53RK
- C20ORF64
- DJ101A2.2
- EC 2.7.11.1
- Kinase PRPK
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
Bud32
SubFamily:
NA
Structure
Mol. Mass (Da):
28,160
# Amino Acids:
253
# mRNA Isoforms:
1
mRNA Isoforms:
28,160 Da (253 AA; Q96S44)
4D Structure:
Interacts with TPRKB.
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 33 | 253 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S34, S140, S250+.
Threonine phosphorylated:
T7, T8, T135.
Tyrosine phosphorylated:
Y201+.
Ubiquitinated:
K40, K134.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 47
47
1776
15
1168
 14
14
546
7
954
 -
-
-
-
-
 18
18
698
69
1791
 21
21
778
23
592
 1.3
1.3
51
18
18
 0.9
0.9
34
23
14
 94
94
3574
22
4545
 0.8
0.8
31
3
2
 2
2
77
59
185
 3
3
131
11
229
 22
22
838
23
680
 10
10
365
2
182
 13
13
506
5
790
 4
4
136
11
267
 5
5
177
12
256
 2
2
85
146
83
 10
10
361
5
457
 3
3
117
41
408
 26
26
968
61
635
 4
4
134
11
235
 1.3
1.3
49
9
12
 -
-
-
-
-
 19
19
705
7
971
 4
4
158
11
229
 43
43
1647
48
2067
 9
9
340
5
424
 16
16
590
5
776
 9
9
336
5
392
 2
2
65
28
38
 44
44
1652
12
24
 100
100
3788
18
7507
 0.2
0.2
8
24
6
 24
24
893
52
702
 2
2
59
35
59
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.5
99.5
100
100 68.3
68.3
71.9
- -
-
-
92 -
-
-
91 78.4
78.4
81.1
94 -
-
-
- 83.7
83.7
88.9
88 -
-
-
88 -
-
-
- -
-
-
- 56.3
56.3
66.2
73 66
66
78.2
73 61.2
61.2
77.4
68 -
-
-
- 41.5
41.5
56.9
47 -
-
-
- -
-
-
45.5 -
-
-
- 44.2
44.2
60.4
- 41.9
41.9
58.1
- -
-
-
47 -
-
-
49 32.1
32.1
48.6
40 -
-
-
53
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TP53 - P04637 |
| 2 | TPRKB - Q9Y3C4 |
| 3 | OSGEP - Q9NPF4 |
| 4 | NAT10 - Q9H0A0 |
| 5 | IMPDH2 - P12268 |
| 6 | PSMD11 - O00231 |
| 7 | NSF - P46459 |
Regulation
Activation:
Protein kinase that phosphorylates 'Ser-15' of p53/TP53 protein and may therefore participate in its activation.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
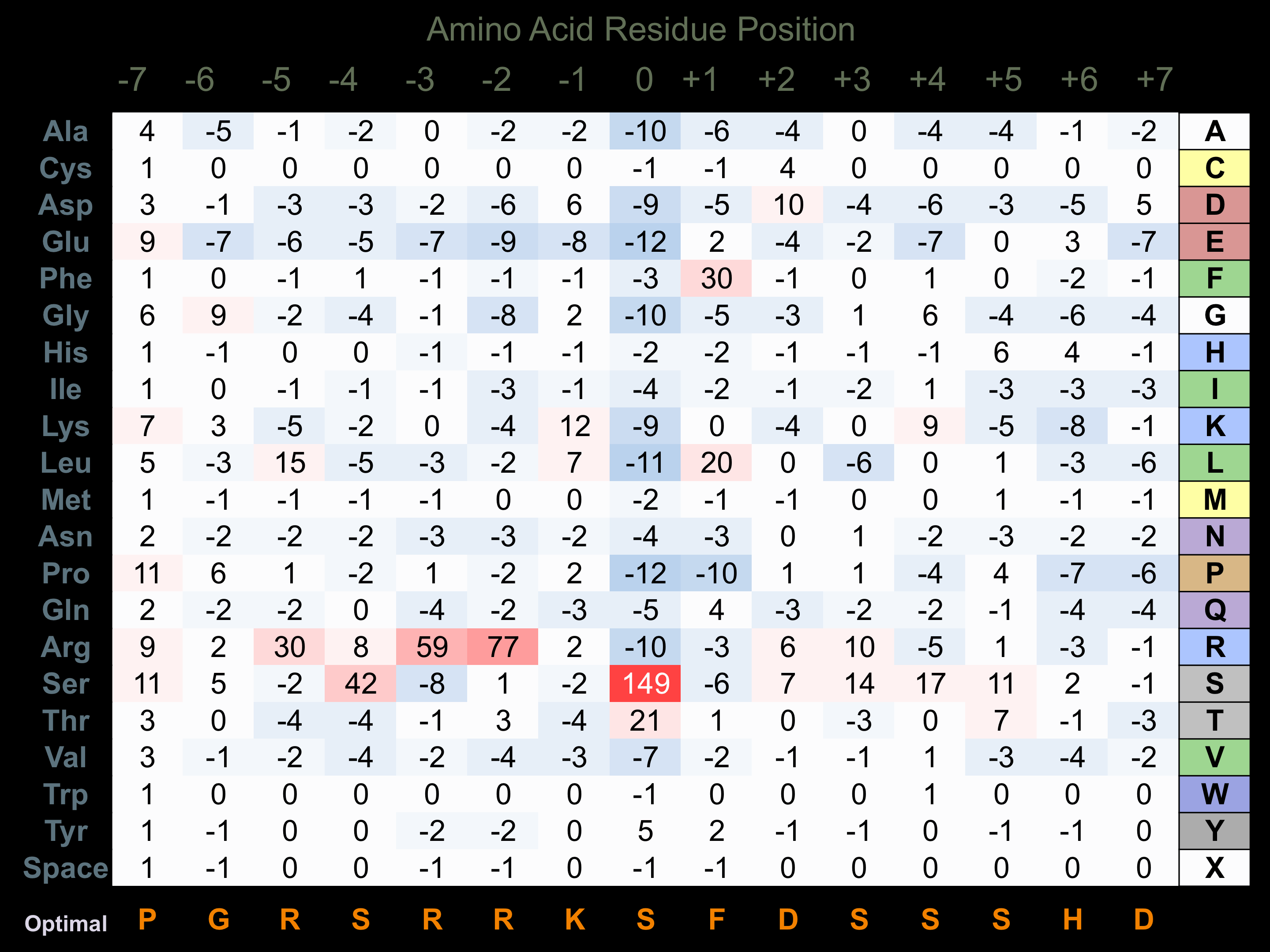
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Eye disorder
Specific Diseases (Non-cancerous):
Shipyard Eye (EKC)
Comments:
Shipyard Eye (EKC) has been related to the disorders St. Louis Encephalitis and Encephalitis. Encephalitis is characterized by inflammation in the brain with symptoms ranging from flu-like to headache, fever, and vomiting. EKC is associated with the Notch and Notch2 signalling pathways. PRPK has also been suggested as a p53 mediator (by phosphorylation), therefore controlling cell cycle and apoptosis but this requires more evidence.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Brain glioblastomas (%CFC= -59, p<0.025). The COSMIC website notes an up-regulated expression score for PRPK in diverse human cancers of 1093, which is 2.4-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 47 for this protein kinase in human cancers was 0.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24726 diverse cancer specimens. This rate is only -24 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.47 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,009 cancer specimens
Comments:
Only 6 deletions (all at L174fs*4), 3 insertions, and no complex mutations are noted on the COSMIC website.

