Nomenclature
Short Name:
IRAK4
Full Name:
Interleukin-1 receptor-associated kinase-4
Alias:
- EC 2.7.11.1
- NY-REN-64
- Interleukin-1 receptor associated kinase 4
- IRAK-4
- IPD1
- REN64
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
IRAK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
51,530
# Amino Acids:
460
# mRNA Isoforms:
2
mRNA Isoforms:
51,530 Da (460 AA; Q9NWZ3); 37,674 Da (336 AA; Q9NWZ3-2)
4D Structure:
IL-1 stimulation leads to the formation of a signaling complex which dissociates from the IL-1 receptor following the binding of PELI1. Interacts with IL1RL1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
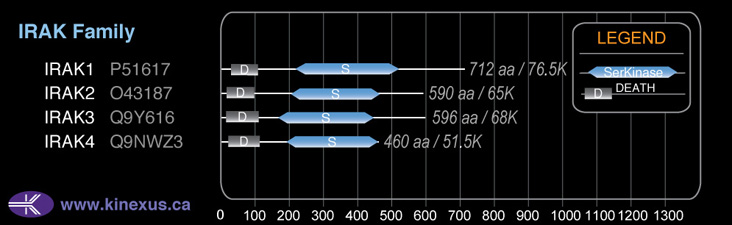
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K3 (N6), K34 (N6).
Serine phosphorylated:
S8, S114, S150, S151, S152, S157, S336, S346+.
Threonine phosphorylated:
T6, T208, T209, T342+, T345+, T352.
Tyrosine phosphorylated:
Y354, Y430.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 23
23
875
16
1092
 0.5
0.5
18
8
15
 1
1
37
1
0
 6
6
236
40
403
 18
18
681
14
681
 1
1
40
37
11
 1
1
40
15
17
 11
11
444
15
528
 12
12
454
10
452
 0.9
0.9
33
33
13
 1.2
1.2
48
12
37
 18
18
696
74
560
 2
2
81
12
15
 0.7
0.7
28
6
17
 2
2
61
9
46
 0.9
0.9
36
7
10
 1.3
1.3
52
15
33
 1
1
37
6
29
 1.2
1.2
47
37
26
 15
15
576
56
624
 0.8
0.8
30
8
28
 2
2
62
10
37
 2
2
72
2
34
 0.7
0.7
29
6
25
 2
2
83
8
83
 16
16
608
22
623
 2
2
66
15
30
 0.7
0.7
26
6
24
 0.6
0.6
25
6
22
 4
4
149
14
70
 26
26
1000
18
662
 100
100
3876
19
7176
 3
3
100
51
288
 23
23
903
26
740
 3
3
104
22
76
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.1
99.1
99.6
99 98.7
98.7
99.1
99 -
-
-
90 -
-
-
95 92.8
92.8
96.3
93 -
-
-
- 83.9
83.9
90.9
84 84.4
84.4
92
84.5 -
-
-
- 45.2
45.2
52
- 47.4
47.4
58.3
67 -
-
-
63 39.1
39.1
54.8
48 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 33.9
33.9
51.1
- 28.1
28.1
47.5
- -
-
-
- -
-
-
39 21.6
21.6
34.4
45 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PELI2 - Q9HAT8 |
| 2 | IL1RL1 - Q01638 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| IRAK4 | Q9NWZ3 | T6 | __MNKPITPSTYVRC | |
| IRAK4 | Q9NWZ3 | S8 | MNKPITPSTYVRCLN | |
| IRAK4 | Q9NWZ3 | S114 | KTANTLPSKEAITVQ | |
| IRAK4 | Q9NWZ3 | S150 | QSYMPPDSSSPENKS | |
| IRAK4 | Q9NWZ3 | S151 | SYMPPDSSSPENKSL | |
| IRAK4 | Q9NWZ3 | S152 | YMPPDSSSPENKSLE | |
| IRAK4 | Q9NWZ3 | T208 | YKGYVNNTTVAVKKL | |
| IRAK4 | Q9NWZ3 | T209 | KGYVNNTTVAVKKLA | |
| IRAK4 | Q9NWZ3 | T342 | ASEKFAQTVMTSRIV | + |
| IRAK4 | Q9NWZ3 | T345 | KFAQTVMTSRIVGTT | + |
| IRAK4 | Q9NWZ3 | S346 | FAQTVMTSRIVGTTA | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| IRAK1 | P51617 | S376 | GSSPSQSSMVARTQT | + |
| IRAK1 | P51617 | T209 | LCEISRGTHNFSEEL | + |
| IRAK1 | P51617 | T387 | RTQTVRGTLAYLPEE | + |
| IRAK4 | Q9NWZ3 | S114 | KTANTLPSKEAITVQ | |
| IRAK4 | Q9NWZ3 | S150 | QSYMPPDSSSPENKS | |
| IRAK4 | Q9NWZ3 | S151 | SYMPPDSSSPENKSL | |
| IRAK4 | Q9NWZ3 | S152 | YMPPDSSSPENKSLE | |
| IRAK4 | Q9NWZ3 | S346 | FAQTVMTSRIVGTTA | + |
| IRAK4 | Q9NWZ3 | S8 | MNKPITPSTYVRCLN | |
| IRAK4 | Q9NWZ3 | T208 | YKGYVNNTTVAVKKL | |
| IRAK4 | Q9NWZ3 | T209 | KGYVNNTTVAVKKLA | |
| IRAK4 | Q9NWZ3 | T342 | ASEKFAQTVMTSRIV | + |
| IRAK4 | Q9NWZ3 | T345 | KFAQTVMTSRIVGTT | + |
| IRAK4 | Q9NWZ3 | T6 | __MNKPITPSTYVRC | |
| p47phox | P14598 | S320 | QRSRKRLSQDAYRRN | ? |
| p47phox | P14598 | S345 | QARPGPQSPGSPLEE | + |
| p47phox | P14598 | S348 | PGPQSPGSPLEEERQ | |
| p47phox | P14598 | S359 | EERQTQRSKPQPAVP | + |
| p47phox | P14598 | S370 | PAVPPRPSADLILNR | + |
| PELI1 | Q96FA3 | S293 | FNTLAFPSMKRKDVV | + |
| PELI1 | Q96FA3 | S70 | PQAAKAISNKDQHSI | |
| PELI1 | Q96FA3 | S76 | ISNKDQHSISYTLSR | + |
| PELI1 | Q96FA3 | S78 | NKDQHSISYTLSRAQ | + |
| PELI1 | Q96FA3 | S82 | HSISYTLSRAQTVVV | + |
| PELI1 | Q96FA3 | T127 | GSQSNSDTQSVQSTI | |
| PELI1 | Q96FA3 | T288 | QCPVGFNTLAFPSMK | + |
| PELI1 | Q96FA3 | T80 | DQHSISYTLSRAQTV | + |
| PELI1 | Q96FA3 | T86 | YTLSRAQTVVVEYTH | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
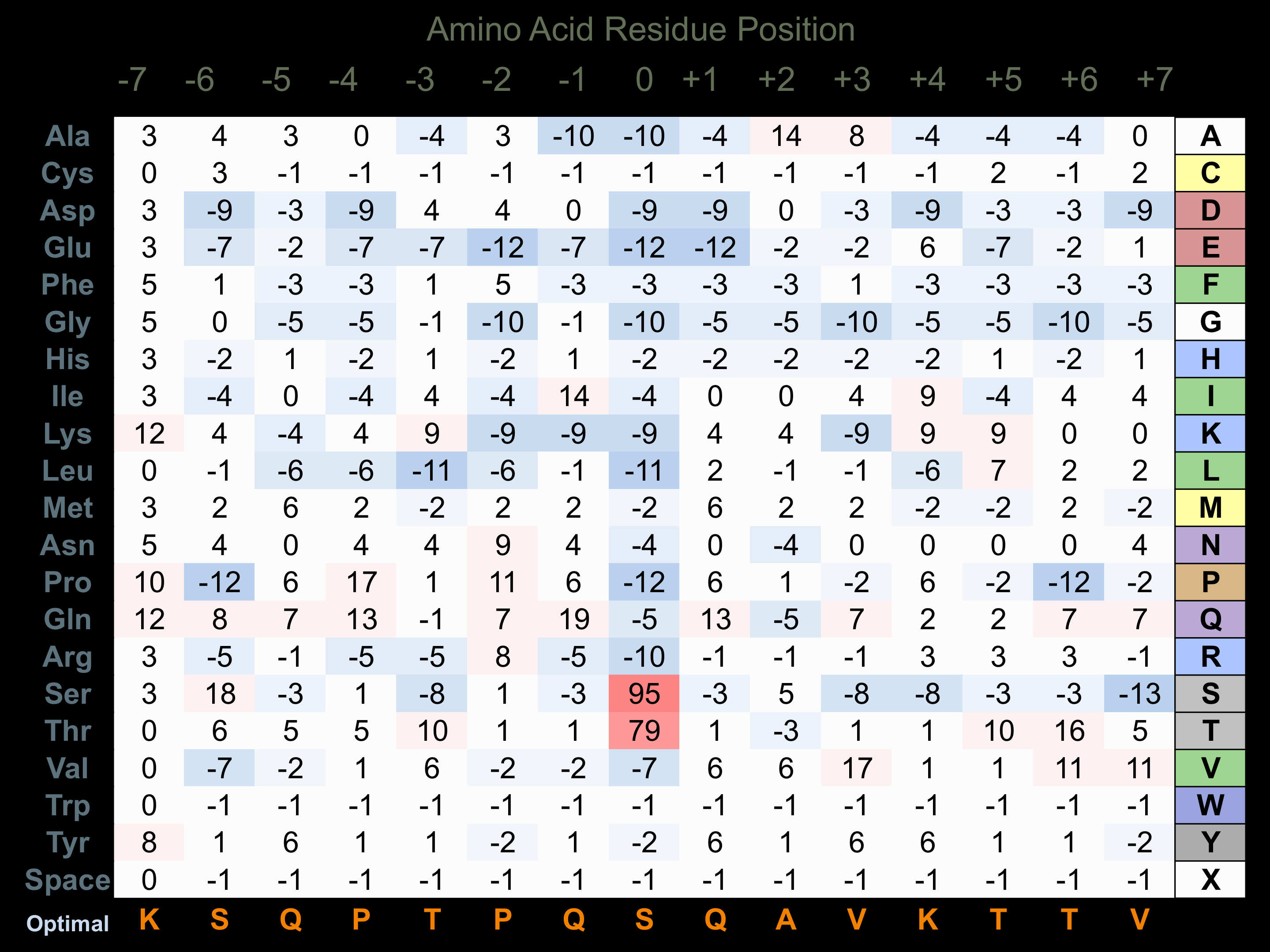
Matrix Type:
Experimentally derived from alignment of 32 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Immune disorders
Specific Diseases (Non-cancerous):
Irak-4 deficiency; Pneumococcal meningitis; Invasive pneumococcal disease, Recurrent isolated, 1
Comments:
IRAK4 deficiency is a rare immunodeficiency condition resulting in severe recurring infections with pyogenic bacteria (inducing pus production), especially Streptococcus pneumoniae. IRAK4 deficiency is linked to TLR signalling and NF-kappaB pathways.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for IRAK4 in diverse human cancers of 397, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 125 for this protein kinase in human cancers was 2.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 25335 diverse cancer specimens. This rate is only -30 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.29 % in 603 endometrium cancers tested; 0.21 % in 1270 large intestine cancers tested; 0.2 % in 864 skin cancers tested; 0.18 % in 238 bone cancers tested; 0.11 % in 575 stomach cancers tested; 0.08 % in 558 thyroid cancers tested; 0.08 % in 548 urinary tract cancers tested; 0.07 % in 1512 liver cancers tested; 0.06 % in 710 oesophagus cancers tested; 0.06 % in 1956 lung cancers tested; 0.04 % in 1490 breast cancers tested; 0.03 % in 2060 central nervous system cancers tested; 0.03 % in 2009 haematopoietic and lymphoid cancers tested; 0.02 % in 939 prostate cancers tested; 0.02 % in 891 ovary cancers tested; 0.02 % in 1276 kidney cancers tested; 0 % in 958 upper aerodigestive tract cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R12C (3).
Comments:
Only 5 deletions, and no insertions or complex mutations are noted on the COSMIC website.

