Nomenclature
Short Name:
ROCK2
Full Name:
Rho-associated protein kinase 2
Alias:
- EC 2.7.11.1
- KIAA0619
- Rho-associated, coiled-coil containing protein kinase 2
- ROK-alpha
- Kinase ROCK2
- P164 ROCK-2
- RhoA-binding serine/threonine kinase alpha
- Rho-associated protein kinase 2
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
DMPK
SubFamily:
ROCK
Specific Links
Structure
Mol. Mass (Da):
160900
# Amino Acids:
1388
# mRNA Isoforms:
1
mRNA Isoforms:
160,900 Da (1388 AA; O75116)
4D Structure:
Homodimer. Binds RHOA that has been activated by GTP binding. Binds IRS1, RHOB and RHOC
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
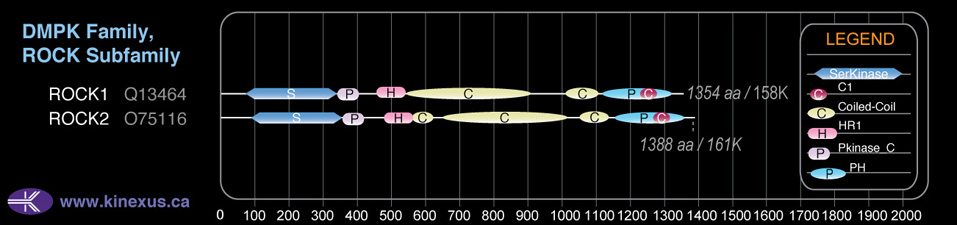
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 92 | 354 | Pkinase |
| 357 | 425 | Pkinase_C |
| 475 | 559 | HR1 |
| 560 | 619 | Coiled-coil |
| 650 | 1015 | Coiled-coil |
| 1054 | 1128 | Coiled-coil |
| 1150 | 1349 | PH |
| 1261 | 1315 | C1 |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K1350, K1351.
Methylated:
K1368.
Serine phosphorylated:
S2, S298, S425, S575, S724, S930, S1099+, S1132, S1133+, S1134, S1137, S1194, S1238, S1361, S1362, S1366, S1374, S1379.
Threonine phosphorylated:
T16, T253+, T414, T567, T573, T577+, T967+, T1078, T1165+, T1212, T1334, T1365.
Tyrosine phosphorylated:
Y722-, Y936, Y1232, Y1318, Y1319.
Ubiquitinated:
K121, K216, K989, K995, K1071.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
938
29
1034
 8
8
74
15
105
 76
76
710
32
446
 55
55
512
115
509
 96
96
898
23
676
 9
9
81
74
46
 40
40
375
31
520
 75
75
702
64
1129
 80
80
752
17
619
 18
18
172
103
147
 19
19
177
49
132
 81
81
761
183
697
 17
17
157
54
117
 11
11
105
12
64
 11
11
101
15
72
 8
8
72
15
37
 33
33
314
140
2075
 40
40
379
41
304
 46
46
433
107
431
 68
68
636
109
624
 21
21
201
42
144
 27
27
256
46
191
 29
29
269
34
170
 17
17
156
41
118
 12
12
113
43
80
 89
89
836
81
1404
 15
15
140
56
123
 21
21
196
40
128
 46
46
433
41
342
 12
12
115
28
127
 77
77
720
24
671
 62
62
578
36
611
 55
55
518
68
781
 87
87
817
52
638
 12
12
115
35
119
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 43.3
43.3
56.9
100 90.4
90.4
90.8
99 -
-
-
98 -
-
-
100 87.8
87.8
89.5
96 -
-
-
- 96.6
96.6
98.3
97 95.7
95.7
97.4
97 -
-
-
- 82.3
82.3
86.2
- 86.8
86.8
91.9
91.5 82
82
89.9
83 75.2
75.2
86.6
77 -
-
-
- 24.1
24.1
45.2
46 -
-
-
- 34.2
34.2
52.8
44 47.4
47.4
64.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | RND3 - P61587 |
| 2 | RHOC - P08134 |
| 3 | LIMK2 - P53671 |
| 4 | MSN - P26038 |
| 5 | RDX - P35241 |
| 6 | PFN2 - P35080 |
| 7 | CDC25A - P30304 |
| 8 | PPP1R12A - O14974 |
| 9 | PTK2 - Q05397 |
| 10 | PPP1R14B - Q96C90 |
| 11 | ADD1 - P35611 |
| 12 | NFKBIA - P25963 |
| 13 | IRS1 - P35568 |
| 14 | VIM - P08670 |
| 15 | CASP3 - P42574 |
Regulation
Activation:
Activated binding to th small G protein RhoA. Activated by phosphorylation at Thr-967 and Ser-1099.
Inhibition:
Inhibited by phosphorylation at Tyr-722.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CRMP2 (DPYSL2) | Q16555 | T555 | DNIPRRTTQRIVAPP | |
| Endophilin 1 | Q99962 | T14 | KKQFHKATQKVSEKV | |
| Ezrin | P15311 | T567 | QGRDKYKTLRQIRQG | |
| IRS1 | P35568 | S636 | SGDYMPMSPKSVSAP | - |
| IRS1 | P35568 | S639 | YMPMSPKSVSAPQQI | - |
| LIMK2 | P53671 | T505 | NDRKKRYTVVGNPYW | + |
| Moesin | P26038 | T558 | LGRDKYKTLRQIRQG | |
| MRLC1 (MYL9) | P24844 | S20 | KRPQRATSNVFAMFD | |
| MRLC2 (MYL12B) | P19105 | S20 | KRPQRATSNVFAMFD | |
| MYPT1 | O14974 | S852 | RPREKRRSTGVSFWT | |
| MYPT1 | O14974 | T696 | ARQSRRSTQGVTLTD | |
| MYPT1 | O14974 | T853 | PREKRRSTGVSFWTQ | |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PPP1R14B | Q96C90 | T57 | VRRQGKVTVKYDRKE | - |
| Radixin | P35241 | T564 | AGRDKYKTLRQIRQG | |
| ROCK2 | O75116 | T414 | QLPFIGFTYYRENLL | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TNNI3 | P19429 | T142 | RGKFKRPTLRRVRIS | |
| Vimentin | P08670 | S39 | TTSTRTYSLGSALRP | - |
| Vimentin | P08670 | S72 | SSAVRLRSSVPGVRL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
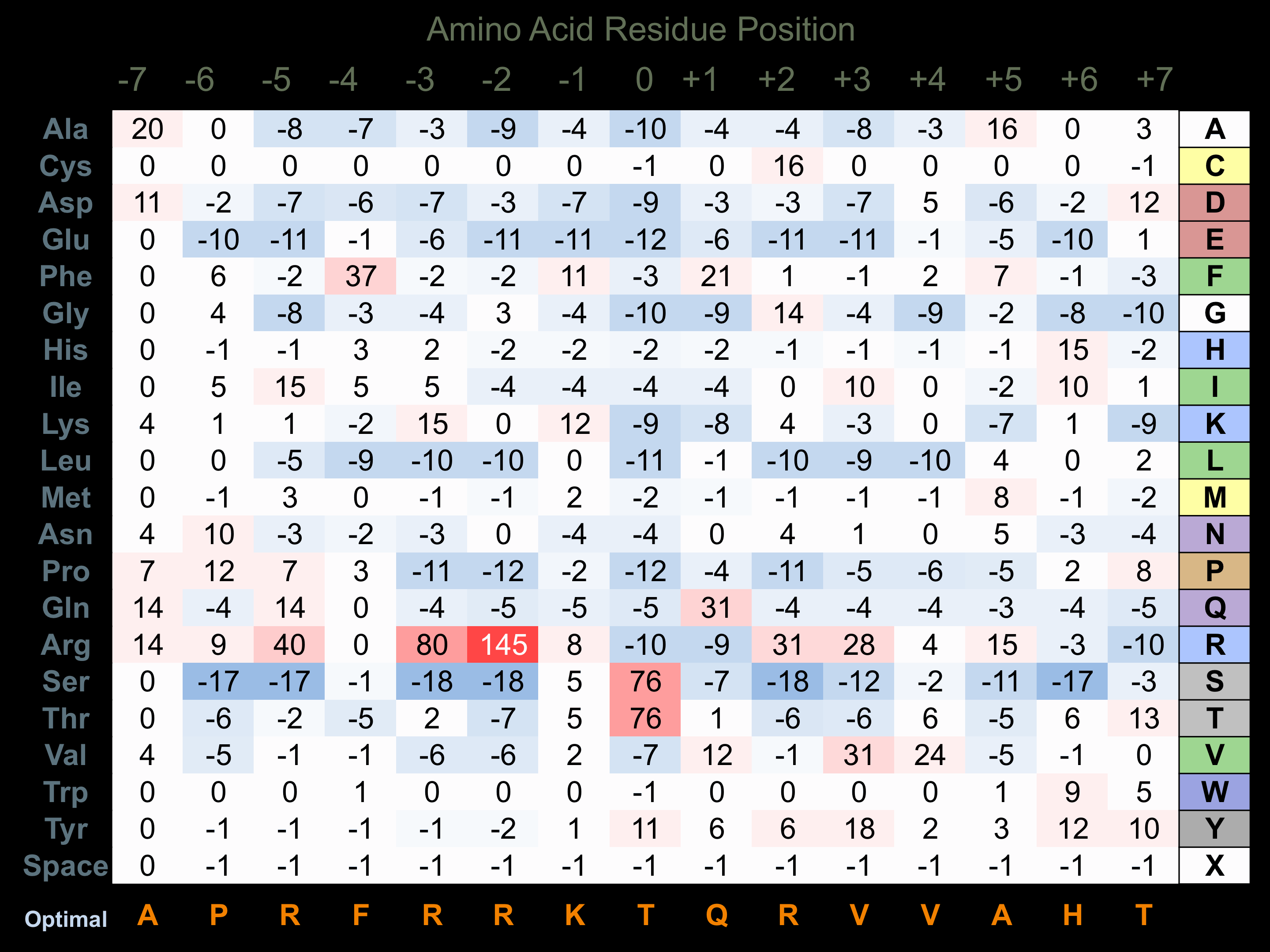
Matrix Type:
Experimentally derived from alignment of 22 known protein substrate phosphosites and 11 peptides phosphorylated by recombinant ROCK2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Immune disorder
Specific Diseases (Non-cancerous):
Behcet's disease
Comments:
Behcet's disease, also known as silk road disease, is a rare immune disease characterized by small blood vessel vasculitis, mucous membrane ulceration, and ocular defects. Affected systems include the gastrointestinal tract, pulmonary, musculoskeletal, cardiovascular, and nervous system. This disease is often fatal due to aneurysms or severe neurological deficits. Polymorphisms in the ROCK2 gene have been associated with patients with Behcet's disease. In addition, patients with Behcet's disease displayed significantly elevated ROCK2 mRNA expression in peripheral blood samples, indicating a potential association between elevated ROCK2 expression and the disease pathology. Therefore, ROCK2 may be a susceptibility gene for the development of Behcet's disease.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Breast epithelial carcinomas (%CFC= +65, p<0.093). The COSMIC website notes an up-regulated expression score for ROCK2 in diverse human cancers of 406, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 20 for this protein kinase in human cancers was 0.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24939 diverse cancer specimens. This rate is only -18 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.28 % in 1270 large intestine cancers tested; 0.27 % in 864 skin cancers tested; 0.19 % in 603 endometrium cancers tested; 0.17 % in 589 stomach cancers tested; 0.17 % in 548 urinary tract cancers tested; 0.11 % in 273 cervix cancers tested; 0.08 % in 1634 lung cancers tested; 0.07 % in 710 oesophagus cancers tested; 0.06 % in 238 bone cancers tested; 0.06 % in 1512 liver cancers tested; 0.05 % in 1316 breast cancers tested; 0.05 % in 1276 kidney cancers tested; 0.03 % in 881 prostate cancers tested; 0.03 % in 833 ovary cancers tested; 0.02 % in 942 upper aerodigestive tract cancers tested; 0.02 % in 441 autonomic ganglia cancers tested; 0.02 % in 382 soft tissue cancers tested; 0.02 % in 2103 central nervous system cancers tested; 0.02 % in 1459 pancreas cancers tested; 0.01 % in 558 thyroid cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R339Q (4); D854V (3).
Comments:
Fourteen deletions (8 at W138fs*31), 7 insertions (3 at W138fs*12), and no complex mutations are noted on the COSMIC website.

