Nomenclature
Short Name:
PLK1
Full Name:
Serine-threonine-protein kinase PLK1
Alias:
- ADRB2
- Polo-like kinase 1
- Serine- threonine protein kinase 13
- STPK13
- EC 2.7.11.21
- Kinase PLK1
- PLK
- PLK-1
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
PLK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
68,255
# Amino Acids:
603
# mRNA Isoforms:
1
mRNA Isoforms:
68,255 Da (603 AA; P53350)
4D Structure:
Interacts with CEP170 and EVI5. Interacts and phosphorylates ERCC6L. Interacts with FAM29A. Interacts with BTBD12/SLX4 and TTDN1. Interacts with BUB1B. Interacts (via POLO-box domain) with the phosphorylated form of BUB1, MLF1IP and CDC25C. Interacts with isoform 3 of SGOL1. Interacts with BORA, KIF2A and AURKA. Interacts with TOPORS and CYLD
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
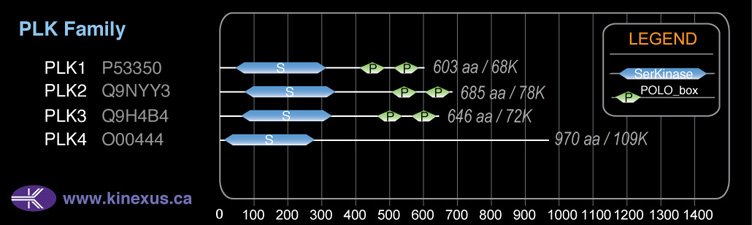
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K9.
Serine phosphorylated:
S2, S49+, S103, S137+, S224, S326+, S330, S331, S335, S375, S383, S387, S450, S461, S466, S565, S578, S597.
Threonine phosphorylated:
T6, T210+, T214-, T341, T366, T459, T464, T498.
Tyrosine phosphorylated:
Y217-, Y309, Y462.
Ubiquitinated:
K38, K146, K200, K272, K338, K474, K480, K492, K589.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 32
32
699
22
1178
 0.9
0.9
21
10
29
 3
3
75
10
104
 68
68
1505
57
4803
 16
16
360
14
366
 4
4
79
43
127
 16
16
350
19
650
 80
80
1765
31
4509
 7
7
152
10
152
 7
7
160
56
281
 4
4
84
23
134
 19
19
420
116
529
 3
3
64
21
90
 2
2
38
9
30
 6
6
123
13
176
 1.1
1.1
24
8
25
 11
11
251
170
302
 3
3
75
17
113
 1.1
1.1
24
52
26
 11
11
247
56
292
 5
5
106
21
157
 5
5
100
23
149
 8
8
180
19
135
 14
14
306
17
297
 15
15
322
21
303
 100
100
2215
37
5592
 5
5
109
24
117
 3
3
64
17
97
 3
3
58
17
79
 7
7
157
14
105
 18
18
407
18
298
 33
33
734
21
655
 75
75
1651
55
1975
 29
29
643
31
646
 8
8
175
22
95
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.8
100 99.8
99.8
99.8
100 -
-
-
96 -
-
-
97 69.6
69.6
71
96 -
-
-
- 95
95
97.7
95 94.2
94.2
96.8
94 -
-
-
- 69.3
69.3
74.8
- 80.1
80.1
89.9
82 79.8
79.8
90.5
82 74.3
74.3
87.6
77 -
-
-
- 50.8
50.8
69.5
55 51.4
51.4
70
- 49.9
49.9
66.3
56 59.4
59.4
76.5
- -
-
-
- 31.8
31.8
50.2
- -
-
-
- 26
26
40.5
- 33.9
33.9
51.6
43 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC25C - P30307 |
| 2 | PIN1 - Q13526 |
| 3 | CHEK2 - O96017 |
| 4 | BRCA2 - P51587 |
| 5 | MYST2 - O95251 |
| 6 | NUDC - Q9Y266 |
| 7 | BUB1 - O43683 |
| 8 | MYT1 - Q01538 |
| 9 | DCTN1 - Q14203 |
| 10 | ECT2 - Q9H8V3 |
| 11 | NPM1 - P06748 |
| 12 | TPT1 - P13693 |
| 13 | PKMYT1 - Q99640 |
| 14 | KIF23 - Q02241 |
| 15 | GORASP1 - Q9BQQ3 |
Regulation
Activation:
Phosphorylation at Ser-49, Ser-137, Thr-210, and Ser-326 increases phosphotransferase activity. Once activated, activity is stimulated by binding target proteins. Binding of target proteins has no effect on the non-activated kinase.
Inhibition:
NA
Synthesis:
By growth-stimulating agents.
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| APC1 | Q9H1A4 | S355 | AALSRAHSPALGVHS | |
| APC1 | Q9H1A4 | S373 | VQRFNISSHNQSPKR | |
| APC1 | Q9H1A4 | S377 | NISSHNQSPKRHSIS | |
| APC1 | Q9H1A4 | S547 | PLSKLLGSLDEVVLL | |
| APC1 | Q9H1A4 | S688 | FDFEGSLSPVIAPKK | |
| APC1 | Q9H1A4 | T291 | KFSEQGGTPQNVATS | |
| APC1 | Q9H1A4 | T520 | GLPAPSLTMSNTMPR | |
| APC1 | Q9H1A4 | T530 | NTMPRPSTPLDGVST | |
| APC1 | Q9H1A4 | T701 | KKARPSETGSDDDWE | |
| APC4 | Q9UJX5 | S779 | KEEVLSESEAENQQA | |
| APC7 | Q9UJX3 | S51 | MAAAGLHSNVRLLSS | |
| APC7 | Q9UJX3 | S57 | HSNVRLLSSLLLTMS | |
| APC7 | Q9UJX3 | S64 | SSLLLTMSNNNPELF | |
| BORA | Q6PGQ7 | S497 | SSNIQMDSGYNTQNC | |
| BORA | Q6PGQ7 | T501 | QMDSGYNTQNCGSNI | |
| BRCA2 | P51587 | S193 | AEVDPDMSWSSSLAT | |
| BRCA2 | P51587 | S205 | LATPPTLSSTVLIVR | |
| BRCA2 | P51587 | S206 | ATPPTLSSTVLIVRN | |
| BRCA2 | P51587 | T203 | SSLATPPTLSSTVLI | |
| BRCA2 | P51587 | T207 | TPPTLSSTVLIVRNE | |
| BubR1 (Bub1B) | O60566 | T1008 | LNANDEATVSVLGEL | + |
| BubR1 (Bub1B) | O60566 | T676 | LSPIIEDSREATHSS | ? |
| BubR1 (Bub1B) | O60566 | T680 | IEDSREATHSSGFSG | ? |
| BubR1 (Bub1B) | O60566 | T792 | PRNSAELTVIKVSSQ | + |
| Cdc16 | Q13042 | S112 | EKYLKDESGFKDPSS | |
| Cdc23 | Q9UJX2 | T556 | QLRNQGETPTTEVPA | |
| Cdc23 | Q9UJX2 | T559 | NQGETPTTEVPAPFF | |
| Cdc25C | P30307 | S198 | SDELMEFSLKDQEAK | |
| Cdc27 | P30260 | S426 | TQPNINDSLEITKLD | |
| Cdc27 | P30260 | S434 | LEITKLDSSIISEGK | |
| Cdc27 | P30260 | S435 | EITKLDSSIISEGKI | |
| Cdc27 | P30260 | T205 | PETVLTETPQDTIEL | |
| Cdc27 | P30260 | T209 | LTETPQDTIELNRLN | |
| Cdc27 | P30260 | T430 | INDSLEITKLDSSII | |
| Cdc27 | P30260 | T446 | EGKISTITPQIQAFN | |
| CEP55 | Q53EZ4 | S436 | PTAALNESLVECPKC | |
| Chk2 (CHEK2) | O96017 | T68 | SSLETVSTQELYSIP | + |
| Claspin | Q9HAW4 | S30 | EADSPSDSGQGSYET | |
| CTNNB1 | P35222 | S718 | QDDPSYRSFHSGGYG | |
| Cyclin B1 | P14635 | S126 | PILVDTASPSPMETS | |
| Cyclin B1 | P14635 | S128 | LVDTASPSPMETSGC | |
| Cyclin B1 | P14635 | S133 | SPSPMETSGCAPAEE | |
| Cyclin B1 | P14635 | S147 | EDLCQAFSDVILAVN | |
| ERCC6L | Q2NKX8 | T1063 | VKQFDASTPKNDISP | |
| FBX5 | Q9UKT4 | S145 | TSRLYEDSGYSSFSL | |
| FBX5 | Q9UKT4 | S149 | YEDSGYSSFSLQSGL | |
| HSF1 | Q00613 | S216 | IPLMLNDSGSAHSMP | |
| HSF1 | Q00613 | S419 | SALLDLFSPSVTVPD | |
| IKKb (IKBKINASE) | O14920 | S733 | TVREQDQSFTALDWS | - |
| IKKb (IKBKINASE) | O14920 | S740 | SFTALDWSWLQTEEE | - |
| IKKb (IKBKINASE) | O14920 | S750 | QTEEEEHSCLEQAS_ | - |
| IRS1 | P35568 | S24 | GYLRKPKSMHKRFFV | |
| Kizuna | Q2M2Z5 | T379 | WSTSSDLTISISEDD | + |
| MAD1L1 | Q9Y6D9 | S22 | RSLNNFISQRVEGGS | |
| MAD1L1 | Q9Y6D9 | S29 | SQRVEGGSGLDISTS | |
| MAD1L1 | Q9Y6D9 | T680 | SKMQLLETEFSHTVG | |
| MgcRacGAP | Q9H0H5 | S149 | RLSTIDESGSILSDI | + |
| MgcRacGAP | Q9H0H5 | S157 | GSILSDISFDKTDES | + |
| MgcRacGAP | Q9H0H5 | S164 | SFDKTDESLDWDSSL | + |
| MgcRacGAP | Q9H0H5 | S170 | ESLDWDSSLVKTFKL | + |
| MgcRacGAP | Q9H0H5 | S214 | AVDQGNESIVAKTTV | + |
| MgcRacGAP | Q9H0H5 | T260 | QPWNSDSTLNSRQLE | |
| MLF1IP | Q71F23 | S77 | TFDPPLHTTAIYADE | |
| MLF1IP | A5D8X7 | T78 | FDPPLHSTAIYADEE | |
| MYST2 | O95251 | S57 | SQSSQDSSPVRNLQS | |
| Myt1 | Q99640 | S426 | CSLLLDSSLSSNWDD | - |
| Myt1 | Q99640 | S435 | SSNWDDDSLGPSLSP | |
| Myt1 | Q99640 | S469 | PRDALDLSDINSEPP | |
| Myt1 | Q99640 | T495 | LLSLFEDTLDPT___ | - |
| Nlp | Q9Y2I6 | S686 | LEELHEKSQEVIWGL | |
| Nlp | Q9Y2I6 | S87 | VRPSDEDSSSLESAA | |
| Nlp | Q9Y2I6 | S88 | RPSDEDSSSLESAAS | |
| Nlp | Q9Y2I6 | T161 | SDEEAESTKEAQNEL | |
| NPM1 | P06748 | S4 | ____MEDSMDMDMSP | |
| NudC | Q9Y266 | S274 | KKINPENSKLSDLDS | |
| NudC | Q9Y266 | S326 | QHPEMDFSKAKFN__ | |
| p73 | O15350 | T27 | SSLEPDSTYFDLPQS | |
| Pin1 | Q13526 | S65 | SHLLVKHSQSRRPSS | |
| PLEKHG6 | Q3KR16 | T574 | HLVVTEDTDEDAPLV | + |
| RACGAP1 | Q9H0H5 | S149 | RLSTIDESGSILSDI | + |
| RACGAP1 | Q9H0H5 | S157 | GSILSDISFDKTDES | + |
| RACGAP1 | Q9H0H5 | S164 | SFDKTDESLDWDSSL | + |
| RACGAP1 | Q9H0H5 | S170 | ESLDWDSSLVKTFKL | + |
| Ran | P62826 | S135 | DRKVKAKSIVFHRKK | |
| ROCK2 | O75116 | S1099 | QAQIAEESQIRIELQ | + |
| ROCK2 | O75116 | S1133 | LHIGLDSSSIGSGPG | + |
| ROCK2 | O75116 | S1374 | MKIQQNQSIRRPSRQ | |
| ROCK2 | O75116 | T967 | ELTEKDATIASLEET | + |
| Separase | Q14674 | S1399 | KVNFSDDSDLEDPVS | |
| Separase | Q14674 | T1363 | AGPHVPFTVFEEVCP | |
| SGO1 | Q5FBB7 | S73 | LALENEKSKVKEAQD | |
| SGO1 | Q5FBB7 | T146 | PQIPLEETELPGQGE | |
| TERF1 | P54274 | S435 | KKLKLISSDSED___ | + |
| TOP2A | P11388 | S1337 | LDSDEDFSDFDEKTD | |
| TOP2A | P11388 | S1525 | PIKYLEESDEDDLF_ | |
| TOP2A | P11388 | T1343 | FSDFDEKTDDEDFVP | |
| TOPORS | Q9NS56 | S718 | KDRDGYESSYRRRTL | |
| TPT1 | P13693 | S46 | TEGNIDDSLIGGNAS | |
| TPT1 | P13693 | S64 | PEGEGTESTVITGVD | |
| Vimentin | P08670 | S83 | GVRLLQDSVDFSLAD | |
| Wee1 | P30291 | S53 | GHSTGEDSAFQEPDS | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
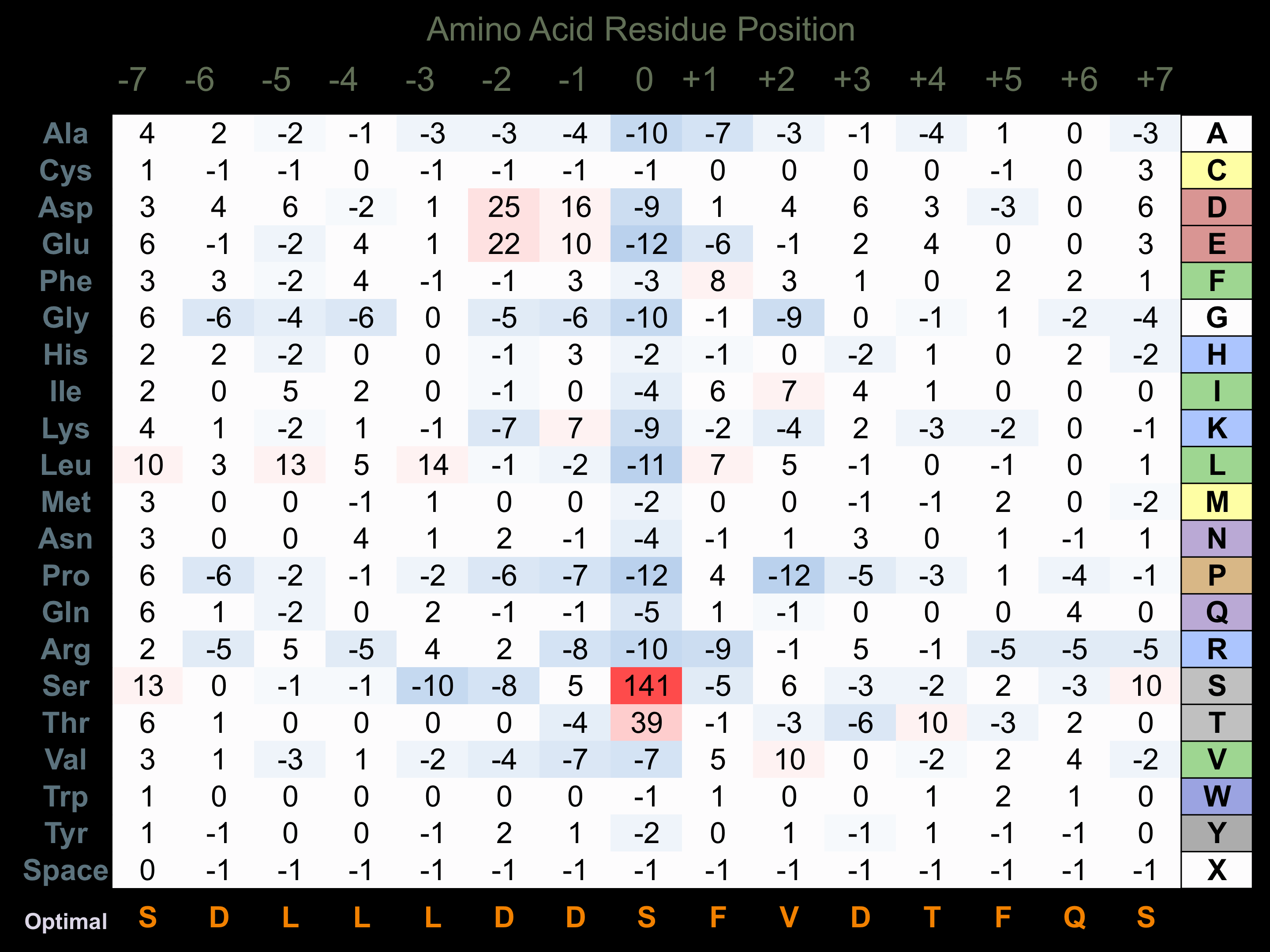
Matrix Type:
Experimentally derived from alignment of 135 known protein substrate phosphosites and 12 peptides phosphorylated by recombinant PLK1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, chromosomal aberration
Specific Diseases (Non-cancerous):
Tetraploidy
Specific Cancer Types:
Gallbladder carcinomas
Comments:
PLK1 might be a tumour requiring protein (TRP). Loss-of-function mutations in Plk1 result in aberrant chromosome segregation during M phase and defects in cytokinesis, leading to chromosomal defects such as polyploidy. Significantly elevated Plk1 expression has been reported in many cancer types, and Plk1 has been suggested as a potential diagnostic biomarker for several tumours. It has been significantly associated with certain clinicopathologic indices in gallbladder carcinomas. The high level expression of Nek7, FoxM1 and Plk1 was significantly associated with shorter overall survival time in 76 specimens of gallbladder carcinomas, pericarcinomas and normal tissues (PubMEd: 23359173).
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +64, p<0.044); Breast sporadic basal-like cancer (BLC) (%CFC= +70, p<0.0001); Cervical cancer (%CFC= -50, p<0.0002); Colon mucosal cell adenomas (%CFC= +166, p<0.0001); Lung adenocarcinomas (%CFC= +229, p<0.0002); Oral squamous cell carcinomas (OSCC) (%CFC= +110, p<0.028); Prostate cancer - primary (%CFC= +103, p<0.02); and Skin melanomas - malignant (%CFC= +48, p<0.004). The COSMIC website notes an up-regulated expression score for PLK1 in diverse human cancers of 556, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 7 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25158 diverse cancer specimens. This rate is very similar (+ 9% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.7 % in 1093 large intestine cancers tested; 0.34 % in 589 stomach cancers tested; 0.25 % in 805 skin cancers tested; 0.13 % in 1753 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S326L (5).
Comments:
Only 5 deletions, 2 insertions and no complex mutations are noted on the COSMIC website.

