Nomenclature
Short Name:
ROR2
Full Name:
Tyrosine-protein kinase transmembrane receptor ROR2
Alias:
- BDB
- BDB1
- EC 2.7.10.1
- MGC163394
- Neurotrophic tyrosine kinase, receptor-related 2
- NTRKR2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Ror
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
104757
# Amino Acids:
943
# mRNA Isoforms:
1
mRNA Isoforms:
104,757 Da (943 AA; Q01974)
4D Structure:
Homodimer; promotes osteogenesis. Binds YWHAB
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
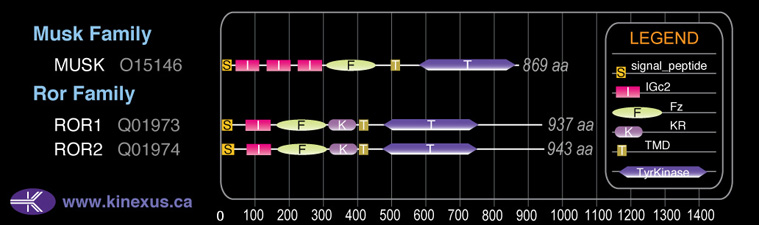
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N70, N188, N318.
O-GalNAcylated:
T157.
Other:
Sulphonated S469, S471.
Serine phosphorylated:
S447, S449, S758, S762, S782.
Threonine phosphorylated:
T438+.
Tyrosine phosphorylated:
Y624, Y645+, Y646+, Y666, Y755, Y824, Y873.
Ubiquitinated:
K489.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 59
59
1651
22
1867
 3
3
80
11
68
 5
5
130
7
213
 31
31
883
87
2578
 17
17
473
23
414
 3
3
83
59
189
 8
8
233
29
555
 51
51
1432
32
2668
 11
11
311
10
277
 2
2
70
75
116
 2
2
52
21
92
 15
15
424
119
489
 1
1
27
18
47
 5
5
130
9
107
 3
3
77
13
116
 1.4
1.4
39
13
44
 16
16
452
114
2140
 3
3
87
14
120
 0.8
0.8
22
67
32
 12
12
341
84
325
 2
2
51
18
66
 2
2
53
20
65
 2
2
65
16
85
 2
2
60
14
56
 2
2
66
18
65
 100
100
2812
59
5070
 1
1
27
19
38
 2
2
67
14
64
 6
6
157
14
150
 5
5
131
28
110
 13
13
364
18
265
 77
77
2170
26
4289
 3
3
78
59
218
 23
23
651
57
645
 3
3
81
35
86
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 95.9
95.9
97
98 95
95
96.7
98 -
-
-
94 -
-
-
- 91.3
91.3
95.1
93 -
-
-
- 92
92
94.7
92.5 92.2
92.2
95.1
92 -
-
-
- 53.5
53.5
67
- 86.1
86.1
92.5
88 79.2
79.2
87.5
82.5 71
71
82.6
73 -
-
-
- 28.5
28.5
44.1
- 30.7
30.7
48
- -
-
-
40 20.4
20.4
36.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | WNT5A - P41221 |
| 2 | FZD2 - Q14332 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
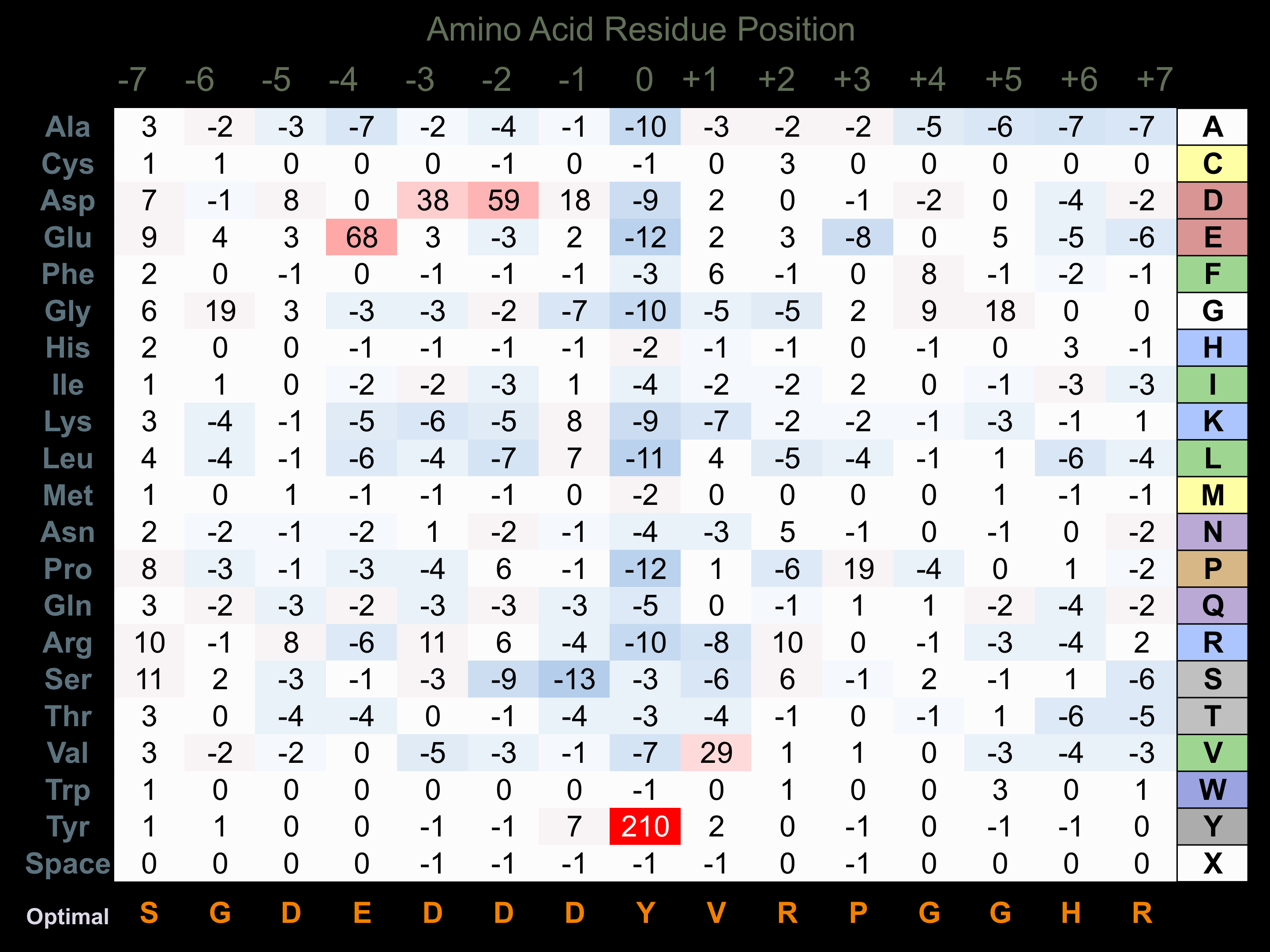
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, genetic disorders
Specific Diseases (Non-cancerous):
Brachydactyly, Type B1; Brachydactyly; Robinow syndrome; Brachydactyly Type B; ROR2-related Robinow syndrome; Robinow syndrome, autosomal recessive; ROR2-related disorders
Comments:
Brachydactyly is a developmental disease characterized by the formation of disproportionately short digits. This disease can occur in isolation or as a component of a more widespread syndrome, and is often accompanied by syndactyly, polydactyly, or reduction defects of the digits. Brachydactyly is inherited in an autosomal dominant manner, showing incomplete penetrance and variable expressivity. Loss-of-function mutations in the ROR2 gene have been observed in patients with brachydactyly, including two truncation mutations (Y755X, W749X) and a frameshift mutation (2249delG leading to a frameshift at gly750 in a arginine/proline-rich sequence). Robinow syndrome is a rare disease characterized by skeletal malformations affecting many parts of the body. Symptoms include shortening of the long bones of the arms and legs, brachydactyly, wedge-shaped vertebrae leading to kyphoscoliosis (abnormal curvature of spine), fused/missing ribs, short stature, and facial abnormalities. Both autosomal dominant and autosomal recessive forms of the disease have been described, distinguished by milder symptoms in the autosomal dominant form. Causal mutations for the autosomal dominant form are unknown, whereas mutations in the ROR2 gene have been observed in patients with the autosomal recessive form of Robinow syndrome, including missense mutations located in both the intracellular and extracellular domains of the ROR2 protein, and a nonsense mutation that produces a truncated protein lacking the tyrosine kinase domain. Therefore, loss-of-function in the ROR2 protein may play an important role in the pathogenesis of these diseases, reflecting an important role for the protein in skeletal development.
Specific Cancer Types:
Nevoid basal cell carcinomas syndrome; Basal cell carcinomas
Comments:
ROR2 appears to be an oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. Gain-of-function mutations in the ROR2 protein are associated specifically in the promotion of the epithelial to mesenchymal transition (EMT), which is a key step during the metastasis of cancer cells. In animal studies, the injection of ROR2 deficient human melanoma cells into a mouse models results in significantly decreased cell motility and spreading as compared to cells that express ROR2, further indicating a role for ROR2 in cancer metastasis. Consequently, ROR2 is suggested to play a role in the promotion of metastasis, specifically in melanoma cells, and contributing to the progression of cancer and acting as an oncoprotein. ROR2 has been implicated in the promotion of metastasis in melanoma cancer cells through interaction with the non-canonical wnt ligand, Wnt5A, which has been shown to increase melanoma cell motility.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial carcinomas (%CFC= -69, p<0.052); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +148, p<0.0002); Prostate cancer (%CFC= +56, p<0.049); and Uterine leiomyomas (%CFC= -59, p<0.004). The COSMIC website notes an up-regulated expression score for ROR2 in diverse human cancers of 368, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 3 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.13 % in 25490 diverse cancer specimens. This rate is 1.7-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.61 % in 1184 large intestine cancers tested; 0.58 % in 854 skin cancers tested; 0.4 % in 589 stomach cancers tested; 0.29 % in 1943 lung cancers tested; 0.19 % in 602 endometrium cancers tested; 0.09 % in 1270 liver cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,774 cancer specimens
Comments:
Only 5 deletions, 1 insertion and 1 complex mutation are noted on the COSMIC website.


