Nomenclature
Short Name:
TGFbR2
Full Name:
TGF-beta receptor type II
Alias:
- AAT3
- FAA3
- TGF-beta type II receptor
- TGFR2
- TGF-beta R2
- TGFbeta-RII; TAAD2
- EC 2.7.11.30
- HNPCC6
- MFS2
- RIIC
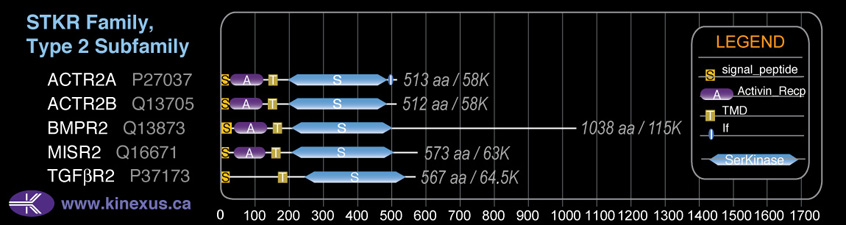
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
STKR
SubFamily:
Type2
Specific Links
Structure
Mol. Mass (Da):
64568
# Amino Acids:
567
# mRNA Isoforms:
2
mRNA Isoforms:
67,457 Da (592 AA; P37173-2); 64,568 Da (567 AA; P37173)
4D Structure:
Binds to DAXX. Interacts with TCTEX1D4
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 23 | signal_peptide |
| 167 | 189 | TMD |
| 244 | 543 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N70, N94N154.
O-GalNAcylated:
T39.
Serine phosphorylated:
S213, S352, S409+, S416-, S548, S553, S562.
Threonine phosphorylated:
T566.
Tyrosine phosphorylated:
Y259+, Y284+, Y336+, Y424-, Y470.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 29
29
1329
26
1023
 6
6
284
17
262
 37
37
1672
8
1302
 9
9
424
91
808
 25
25
1120
22
882
 4
4
167
80
258
 11
11
511
35
640
 16
16
729
40
690
 16
16
710
17
676
 8
8
381
93
515
 6
6
264
31
572
 26
26
1170
181
815
 16
16
725
30
956
 3
3
156
15
146
 8
8
356
25
400
 5
5
237
16
269
 5
5
227
202
1045
 9
9
410
19
608
 5
5
208
89
290
 18
18
802
109
736
 6
6
276
23
405
 17
17
757
27
1013
 19
19
876
18
907
 3
3
150
19
282
 11
11
496
23
556
 18
18
801
55
783
 8
8
354
33
510
 13
13
591
19
843
 18
18
798
19
960
 4
4
169
28
129
 27
27
1242
24
776
 100
100
4557
40
9398
 12
12
531
72
877
 26
26
1204
57
816
 2
2
81
35
100
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 94
94
94
100 98.9
98.9
99.7
99 -
-
-
92 -
-
-
- 89.5
89.5
93.5
94 -
-
-
- 88.2
88.2
92.9
91 91.5
91.5
96.3
90.5 -
-
-
- 74.5
74.5
82.1
- 75
75
85.2
79 34
34
51.7
72 66.8
66.8
79.9
72 -
-
-
- -
-
-
- -
-
-
- 25.1
25.1
42.2
- 32.4
32.4
49.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
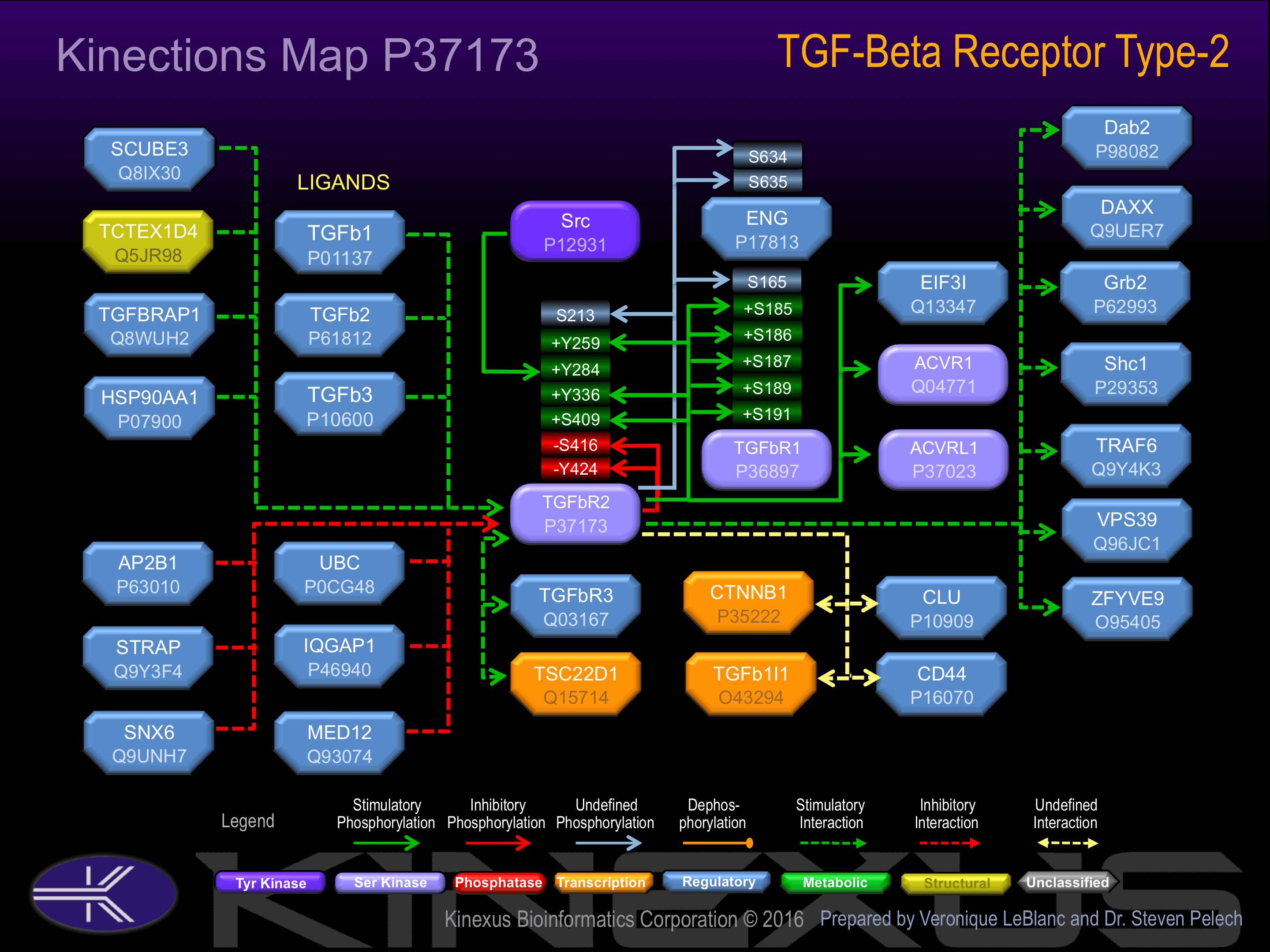
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TGFB3 - P10600 |
| 2 | TGFB1 - P01137 |
| 3 | TGFBR3 - Q03167 |
| 4 | ACVRL1 - P37023 |
| 5 | TTC5 - Q8N0Z6 |
| 6 | AP2B1 - P63010 |
| 7 | CD44 - P16070 |
| 8 | PML - P29590 |
| 9 | CCNB2 - O95067 |
| 10 | SMAD7 - O15105 |
| 11 | SMURF2 - Q9HAU4 |
| 12 | ZFYVE9 - O95405 |
| 13 | DAB2 - P98082 |
| 14 | PIK3R2 - O00459 |
| 15 | PIK3R1 - P27986 |
Regulation
Activation:
Activated by binding transforming growth factor-beta (TGFb), which appears to induce heterodimerization and autophosphorylation. Phosphorylation of Tyr-259, Tyr-336, S409 and Tyr-424 increases phosphotransferase acivity. Phosphorylation at Tyr-284 induces interaction with Grb2 and Shc1.
Protein Info
Short Name:
TGFBR1
Full Name:
Transforming growth factor-beta
Inhibition:
Phosphorylation of Ser-416 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ENG | P17813 | S634 | VAVAAPASSESSSTN | |
| ENG | P17813 | S635 | AVAAPASSESSSTNH | |
| TGFbR1 | P36897 | S165 | VPNEEDPSLDRPFIS | |
| TGFbR1 | P36897 | S187 | LIYDMTTSGSGSGLP | |
| TGFbR1 | P36897 | S189 | YDMTTSGSGSGLPLL | |
| TGFbR1 | P36897 | S191 | MTTSGSGSGLPLLVQ | |
| TGFbR1 | P36897 | T185 | KDLIYDMTTSGSGSG | |
| TGFbR1 | P36897 | T186 | DLIYDMTTSGSGSGL | |
| TGFbR2 | P37173 | S409 | LRLDPTLSVDDLANS | + |
| TGFbR2 | P37173 | S416 | SVDDLANSGQVGTAR | - |
| TGFbR2 | P37173 | Y259 | KGRFAEVYKAKLKQN | + |
| TGFbR2 | P37173 | Y336 | AKGNLQEYLTRHVIS | + |
| TGFbR2 | P37173 | Y424 | GQVGTARYMAPEVLE | - |
| TGFbR2 | P37173 | S213 | TRKLMEFSEHCAIIL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
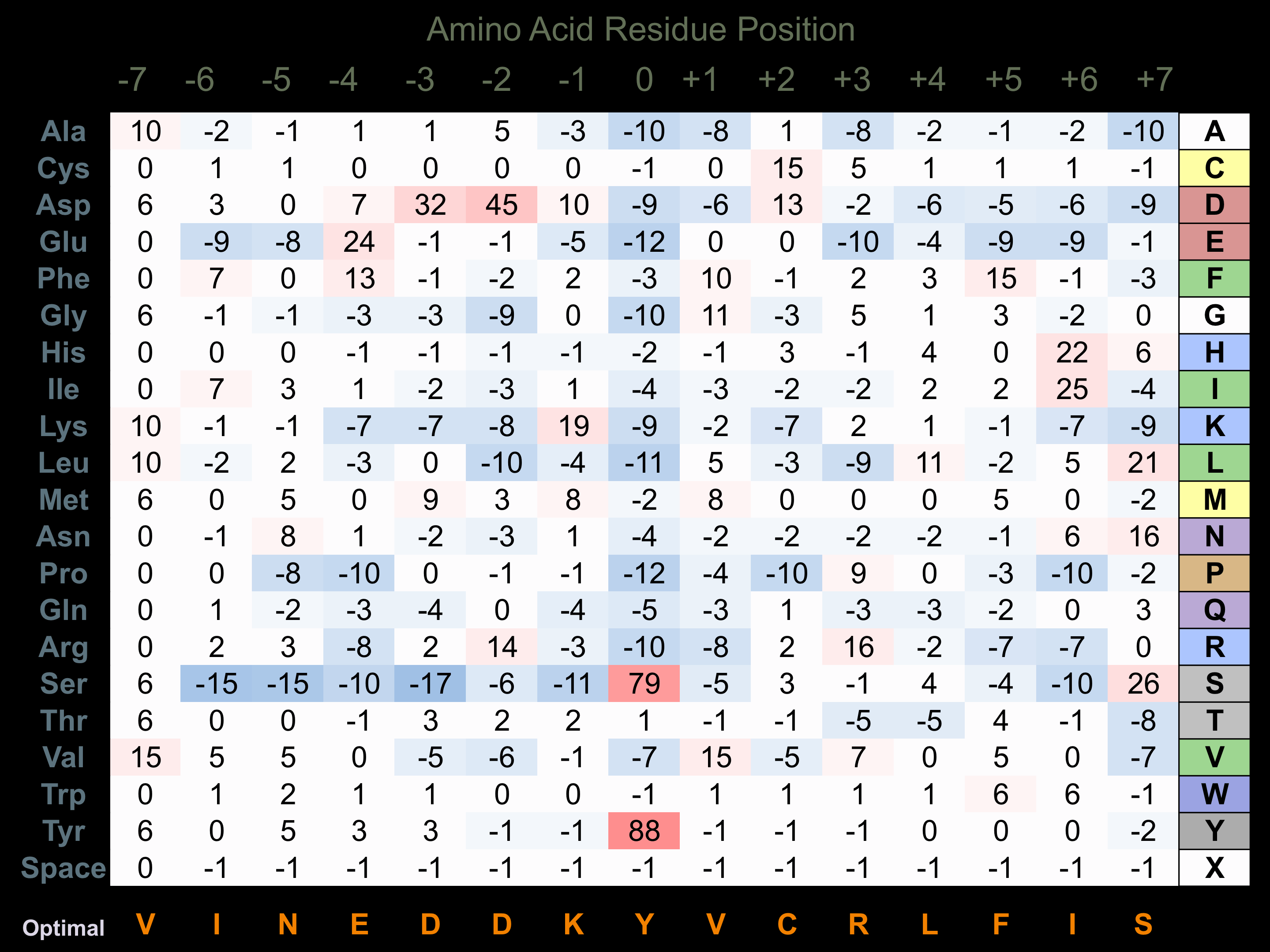
Matrix Type:
Experimentally derived from alignment of 15 known protein substrate phosphosites and 16 peptides phosphorylated by recombinant TGFBR2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Connective tissue disorders
Specific Diseases (Non-cancerous):
Loeys-Dietz syndrome, Type 2; Loeys-Dietz syndrome; Marfan syndrome; Aneurysm; Aortic aneurysm; Thoracic aortic aneurysms and aortic dissections; Intracranial hypotension; Normal pressure hydrocephalus; Kabuki syndrome; Clubfoot; Transient hypogammaglobulinemia; Transient hypogammaglobulinemia of infancy; Chromosome 3p deletion; Loeys-Dietz syndrome Type 1b; Loeys-Dietz syndrome, Type 1; Aortic aneurysm, familial thoracic 4; TGFBR2-related thoracic aortic aneurysms and aortic dissections; TGFBR2-related Loeys-Dietz syndrome
Comments:
TGFBR2 appears to be a tumour suppressor protein (TSP). Cancer-related mutations in human tumours point to a loss of function of the protein kinase.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -93, p<0.068); Classical Hodgkin lymphomas (%CFC= -48, p<0.063); Colon mucosal cell adenomas (%CFC= +50, p<0.014); Colorectal adenocarcinomas (early onset) (%CFC= +84, p<0.076); Lung adenocarcinomas (%CFC= -65, p<0.0001); Ovary adenocarcinomas (%CFC= -73, p<0.003); Skin melanomas - malignant (%CFC= -68, p<0.039); T-cell prolymphocytic leukemia (%CFC= -68, p<0.026); Uterine leiomyomas (%CFC= -79, p<0.0001); and Vulvar intraepithelial neoplasia (%CFC= -52, p<0.085). The COSMIC website notes an up-regulated expression score for TGFBR2 in diverse human cancers of 261, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 54 for this protein kinase in human cancers was 0.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.17 % in 25961 diverse cancer specimens. This rate is 2.2-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.14 % in 1269 large intestine cancers tested; 0.89 % in 636 stomach cancers tested; 0.42 % in 1285 pancreas cancers tested; 0.37 % in 805 skin cancers tested; 0.35 % in 602 endometrium cancers tested; 0.13 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D446N (9); D446V (3); L452P (4); R495* (7); R497* (7); R528H (7); R528C (5).
Comments:
Over 45 deletion mutations (33 at K128fs*35 and 6 at K128fs*3), 9 insertions mutants (7 at P129fs*3) primarily in the extracellular domain of the receptor.