Nomenclature
Short Name:
TRKB
Full Name:
BDNF-NT-3 growth factors receptor
Alias:
- EC 2.7.10.1
- GP145-TrkB
- GP145-TrkB,GP95-TrkB
- NTRK2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Trk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
91,999
# Amino Acids:
822
# mRNA Isoforms:
7
mRNA Isoforms:
93,826 Da (838 AA; Q16620-4); 91,999 Da (822 AA; Q16620); 81,569 Da (735 AA; Q16620-6); 60,994 Da (553 AA; Q16620-5); 59,167 Da (537 AA; Q16620-3); 53,051 Da (477 AA; Q16620-2); 35,332 Da (321 AA; Q16620-7)
4D Structure:
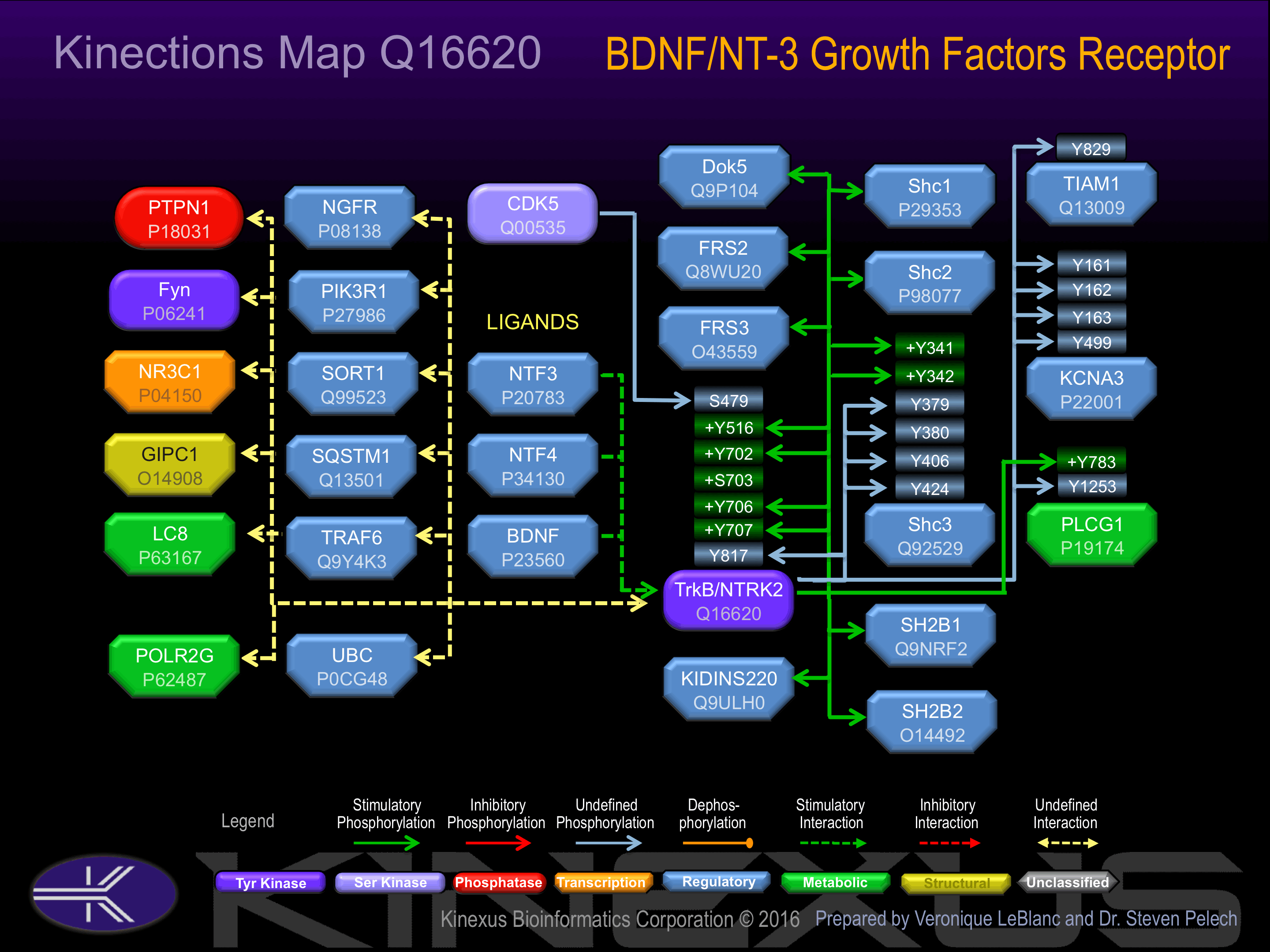
Exists in a dynamic equilibrium between monomeric (low affinity) and dimeric (high affinity) structures. Binds SH2B2. Interacts with SQSTM1 and KIDINS220
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
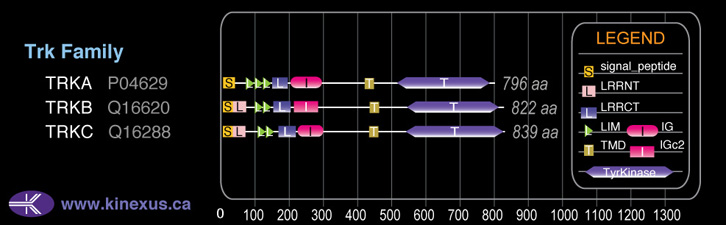
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K461.
N-GlcNAcylated:
N67,N95, N121, N178, N205, N241, N254, N280, N325, N338, N412.
Serine phosphorylated:
S141, S153, S256, S257, S260, S479, S698+, S703+.
Threonine phosphorylated:
T151, T161, T253, T506, T573.
Tyrosine phosphorylated:
Y516+, Y558, Y702+, Y706+, Y707+, Y817.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 85
85
962
67
1149
 3
3
39
33
61
 9
9
100
20
118
 79
79
898
269
993
 81
81
926
62
790
 11
11
125
181
281
 22
22
245
91
437
 34
34
384
100
661
 26
26
291
31
201
 8
8
92
257
108
 3
3
33
75
39
 44
44
503
419
597
 7
7
82
64
76
 4
4
50
29
39
 6
6
72
50
83
 3
3
36
43
49
 2
2
26
860
113
 9
9
98
39
142
 5
5
54
221
59
 48
48
549
271
573
 46
46
529
63
992
 5
5
57
68
79
 3
3
36
48
47
 4
4
49
40
57
 3
3
36
61
47
 68
68
774
162
938
 6
6
66
63
88
 10
10
119
40
149
 7
7
77
39
102
 13
13
153
84
213
 25
25
282
48
280
 100
100
1138
76
2580
 12
12
140
141
468
 57
57
650
171
627
 4
4
46
87
44
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 55.9
55.9
70.7
100 99.9
99.9
99.9
100 -
-
-
92.5 -
-
-
96 96.4
96.4
97.1
98 -
-
-
- 93.9
93.9
97.2
94 93.5
93.5
96.6
94 -
-
-
- -
-
-
- 77.6
77.6
86.5
78 22.3
22.3
35.6
72 55.5
55.5
66.2
65 -
-
-
- 27.5
27.5
45
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NTF4 - P34130 |
| 2 | BDNF - P23560 |
| 3 | NGFR - P08138 |
| 4 | DOK5 - Q9P104 |
| 5 | NCK2 - O43639 |
| 6 | NCK1 - P16333 |
| 7 | PIK3R1 - P27986 |
| 8 | SHC2 - P98077 |
| 9 | DYNLL1 - P63167 |
| 10 | KIDINS220 - Q9ULH0 |
| 11 | RPS27A - P62988 |
| 12 | FRS2 - Q8WU20 |
Regulation
Activation:
Activated by binding BDNF, NT-3, NT-4 or NT-5 growth factor, which induces dimerization and autophosphorylation. Phosphorylation at Tyr-516 increases phosphotransferase activity and induces interaction with PLCg1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Kv1.3 | P22001 | Y161 | PSFDAILYYYQSGGR | |
| Kv1.3 | P22001 | Y162 | SFDAILYYYQSGGRI | |
| Kv1.3 | P22001 | Y163 | FDAILYYYQSGGRIR | |
| Kv1.3 | P22001 | Y499 | EGEEQSQYMHVGSCQ | |
| PLCG1 | P19174 | Y1253 | EGSFESRYQQPFEDF | |
| PLCG1 | P19174 | Y783 | EGRNPGFYVEANPMP | + |
| Shc3 | Q92529 | Y341 | GDGSDHPYYNSIPSK | ? |
| Shc3 | Q92529 | Y342 | DGSDHPYYNSIPSKM | ? |
| Shc3 | Q92529 | Y379 | FAGKEQTYYQGRHLG | |
| Shc3 | Q92529 | Y380 | AGKEQTYYQGRHLGD | |
| Shc3 | P29353 | Y406 | RQGSSDIYSTPEGKL | |
| Shc3 | P29353 | Y424 | PTGEAPTYVNTQQIP | |
| Tiam1 | Q13009 | Y829 | PQPEEDIYELLYKEI | |
| TrkB (NTRK2) | Q16620 | Y516 | PVIENPQYFGITNSQ | + |
| TrkB (NTRK2) | Q16620 | Y702 | FGMSRDVYSTDYYRV | + |
| TrkB (NTRK2) | Q16620 | Y706 | RDVYSTDYYRVGGHT | + |
| TrkB (NTRK2) | Q16620 | Y707 | DVYSTDYYRVGGHTM | + |
| TrkB (NTRK2) | Q16620 | Y817 | LAKASPVYLDILG__ |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
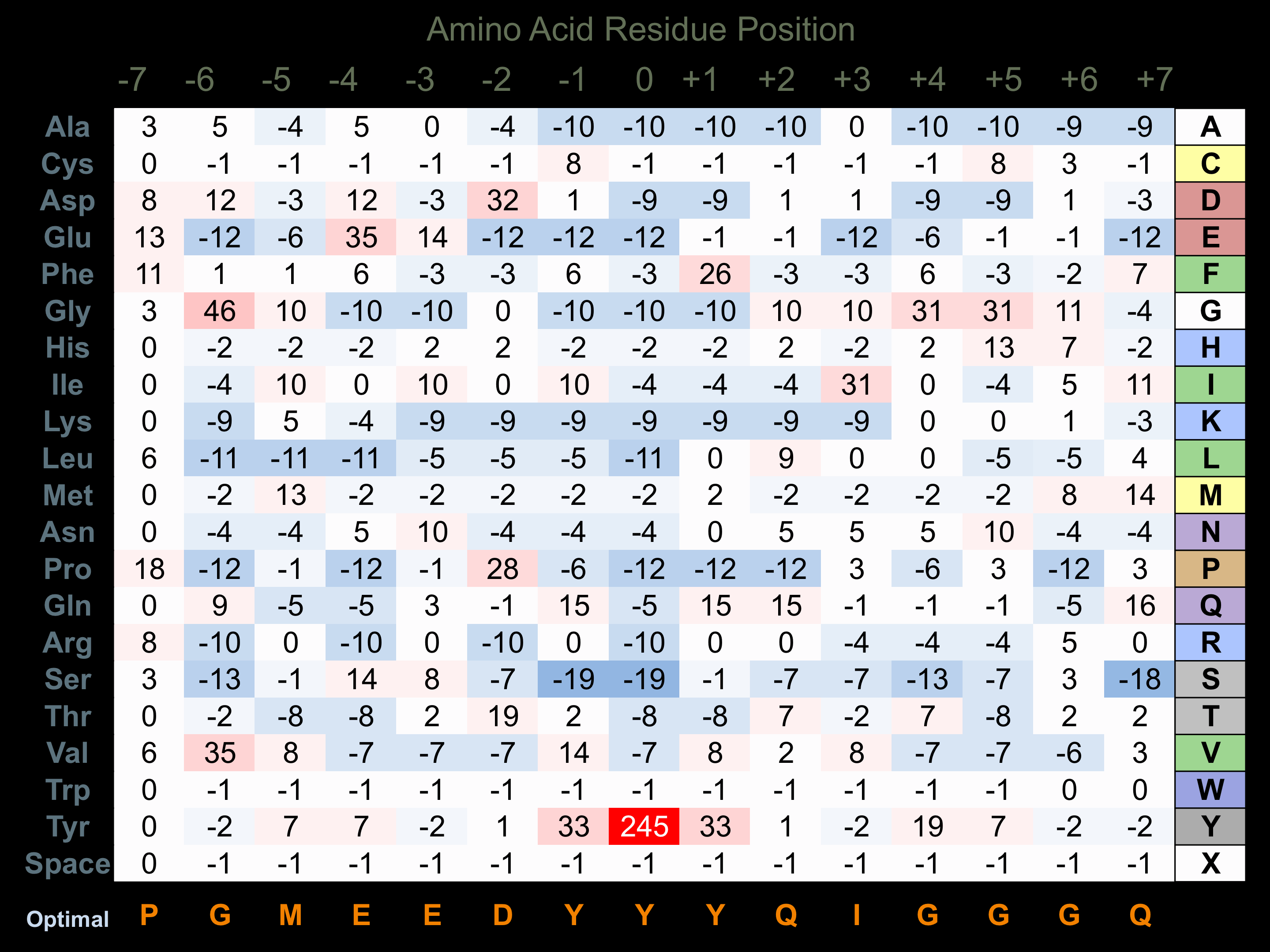
Matrix Type:
Experimentally derived from alignment of 29 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, metabolic disorders
Specific Diseases (Non-cancerous):
Obesity; Obesity, Hyperphagia, and developmental delay (OHPDD)
Comments:
Obesity is considered a rare disease with regard to a genetic propensity. Obesity can arise from excessive weight in muscle, bone, fat, and or body water. Obesity, Hyperphagia, and Developmental Delay (OHPDD) is a condition characterized by obesity at a young age, increased appetite for food (hyperphagia), and a severe delay in the development of speech, language, and motor control. In OHPDD an Y706C mutation in the TRKB gene will inhibit phosphorylation.
Specific Cancer Types:
Pilocytic astrocytomas; Neuroblastomas (NB)
Comments:
TRKB is linked to Pilocytic Astrocytomas, which are benign tumours of the brain or spinal cord that grow slowly.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -48, p<0.002); Breast epithelial carcinomas (%CFC= -53, p<0.048); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -78, p<0.0006); Head and neck squamous cell carcinomas (HNSCC) (%CFC= +81, p<0.033); Lung adenocarcinomas (%CFC= -57, p<0.0001); Malignant pleural mesotheliomas (MPM) tumours (%CFC= -67, p<0.011); Pituitary adenomas (ACTH-secreting) (%CFC= -79, p<0.095); Skin melanomas - malignant (%CFC= -69, p<0.001); and Skin squamous cell carcinomas (%CFC= -52, p<0.032). The COSMIC website notes an up-regulated expression score for TRKB in diverse human cancers of 353, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 1 for this protein kinase in human cancers was 98% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 25532 diverse cancer specimens. This rate is only 37 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.48 % in 1093 large intestine cancers tested; 0.35 % in 805 skin cancers tested; 0.31 % in 589 stomach cancers tested; 0.3 % in 602 endometrium cancers tested; 0.24 % in 1943 lung cancers tested; 0.14 % in 1270 liver cancers tested; 0.1 % in 1488 breast cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,678 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.