Nomenclature
Short Name:
TTK
Full Name:
Dual specificity protein kinase TTK
Alias:
- Cancer/testis antigen 96
- CT96
- Mps1
- MPS1L1
- Phosphotyrosine picked threonine-protein kinase
- PYT; TTK protein kinase
- EC 2.7.12.1
- ESK
- Esk1
- FLJ38280
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
TTK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
97072
# Amino Acids:
857
# mRNA Isoforms:
2
mRNA Isoforms:
97,072 Da (857 AA; P33981); 96,944 Da (856 AA; P33981-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
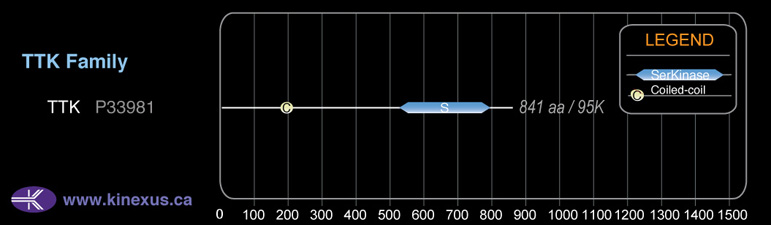
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 161 | 184 | Coiled-coil |
| 525 | 791 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K852, K853.
Methylated:
K19, K385, R467.
Serine phosphorylated:
S3, S7, S15, S37, S80, S108, S214, S216, S218, S258, S281, S291, S317, S321, S333, S345, S353, S362, S363, S382, S393, S433, S436, S440, S455, S464, S582+, S618, S677+, S742+, S821, S824, S837, S845, S847.
Threonine phosphorylated:
T12, T33, T46, T210, T288+, T351, T360, T371, T453, T458, T468, T564, T594, T675-, T676+, T686+, T795, T805, T806-, T849.
Tyrosine phosphorylated:
Y131, Y255, Y374, Y462, Y811+, Y833, Y836.
Ubiquitinated:
K86, K372, K385, K538, K546, K696, K810, K827, K848.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 71
71
1704
16
3599
 0.4
0.4
10
10
7
 8
8
194
22
234
 45
45
1085
67
3881
 13
13
319
14
327
 37
37
888
45
1973
 12
12
297
19
506
 91
91
2198
42
6448
 10
10
242
10
237
 1.1
1.1
27
66
46
 2
2
55
35
100
 13
13
318
125
454
 4
4
105
33
216
 0.7
0.7
16
8
9
 2
2
51
32
109
 0.4
0.4
10
8
7
 1.5
1.5
35
120
118
 6
6
154
29
260
 1.2
1.2
28
64
48
 17
17
404
56
463
 3
3
61
31
76
 4
4
99
32
112
 13
13
309
31
388
 100
100
2413
29
3357
 85
85
2046
31
2840
 89
89
2155
49
6475
 13
13
307
36
496
 4
4
90
29
146
 3
3
75
29
110
 3
3
61
14
37
 18
18
432
18
322
 14
14
328
21
396
 48
48
1151
55
1306
 27
27
644
31
662
 23
23
544
22
844
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.2
99.2
99.7
99 85.8
85.8
87.8
97 -
-
-
87 -
-
-
93 87.4
87.4
92.7
87.5 -
-
-
- 74.3
74.3
84.8
76 76.9
76.9
85.7
79 -
-
-
- 64.3
64.3
73.7
- 61.1
61.1
74.5
64 52.8
52.8
68.5
56 41.5
41.5
57
48.5 -
-
-
- -
-
-
32 -
-
-
- -
-
-
- -
-
-
- 23.1
23.1
40.3
- -
-
-
- -
-
-
- 23
23
42.4
- 22.8
22.8
44.6
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDCA8 - Q53HL2 |
| 2 | TACC2 - O95359 |
| 3 | TUBB1 - Q9H4B7 |
| 4 | AZU1 - P20160 |
| 5 | TUBB2A - Q13885 |
| 6 | TUBB - P07437 |
| 7 | CDC20 - Q12834 |
Regulation
Activation:
Phosphorylation of S582, Thr-675, Thr-676, Thr-686, Ser-742 increases phosphotransferase activity.
Inhibition:
Phosphorylation of Thr-806 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| TTK | P33981 | T33 | KFKNEDLTDELSLNK | |
| TTK | P33981 | S37 | EDLTDELSLNKISAD | |
| TTK | P33981 | S80 | EKNSVPLSDALLNKL | |
| TTK | P33981 | T288 | SPDCDVKTDDSVVPC | ? |
| CHK2 | O96017 | T288 | SPDCDVKTDDSVVPC | + |
| TTK | P33981 | T360 | SITLKNKTESSLLAK | |
| TTK | P33981 | S363 | LKNKTESSLLAKLEE | |
| TTK | P33981 | S436 | VFSVSKQSPPISTSK | |
| TTK | P33981 | T564 | LEEADNQTLDSYRNE | |
| TTK | P33981 | S582 | LNKLQQHSDKIIRLY | + |
| TTK | P33981 | T675 | ANQMQPDTTSVVKDS | + |
| TTK | P33981 | T676 | NQMQPDTTSVVKDSQ | + |
| TTK | P33981 | S677 | QMQPDTTSVVKDSQV | + |
| TTK | P33981 | T686 | VKDSQVGTVNYMPPE | + |
| TTK | P33981 | S742 | QQIINQISKLHAIID | + |
| TTK | P33981 | T795 | HPYVQIQTHPVNQMA | |
| TTK | P33981 | T805 | VNQMAKGTTEEMKYV | |
| TTK | P33981 | T806 | NQMAKGTTEEMKYVL | - |
| TTK | P33981 | Y811 | GTTEEMKYVLGQLVG | + |
| TTK | P33981 | S821 | GQLVGLNSPNSILKA | ? |
| ERK2 | P28482 | S821 | GQLVGLNSPNSILKA | ? |
| ERK1 | P27361 | S821 | GQLVGLNSPNSILKA | ? |
| TTK | P33981 | S824 | VGLNSPNSILKAAKT | |
| TTK | P33981 | S845 | GGESHNSSSSKTFEK | |
| TTK | P33981 | S847 | ESHNSSSSKTFEKKR | |
| TTK | P33981 | T849 | HNSSSSKTFEKKRGK |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Abl1 | P00519 | T735 | DTEWRSVTLPRDLQS | + |
| Abl1 iso2 | P00519-2 | T754 | DTEWRSVTLPRDLQS | + |
| BLM | P54132 | S1290 | QKYSEWTSPAEDSSP | |
| BLM | P54132 | S1296 | TSPAEDSSPGISLSS | |
| BLM | P54132 | S144 | KKLEFSSSPDSLSTI | |
| Borealin | Q96AM3 | T169 | KRSSRANTVTPAVGR | |
| Borealin | Q96AM3 | T230 | DSKEIFLTVPVGGGE | |
| Borealin | Q96AM3 | T88 | QALEEAATADLDITE | |
| Borealin | Q96AM3 | T94 | ATADLDITEINKLTA | |
| BubR1 (Bub1B) | O60566 | S1043 | WKVGKLTSPGALLFQ | |
| BubR1 (Bub1B) | O60566 | S435 | REAELLTSAEKRAEM | |
| BubR1 (Bub1B) | O60566 | S543 | LSEKKNKSPPADPPR | |
| BubR1 (Bub1B) | O60566 | S670 | TLSIKKLSPIIEDSR | ? |
| Chk2 (CHEK2) | O96017 | T68 | SSLETVSTQELYSIP | + |
| HSPA9B | P38646 | S65 | IDLGTTNSCVAVMEG | + |
| HSPA9B | P38646 | T62 | VVGIDLGTTNSCVAV | + |
| Tau iso8 | P10636-8 | S208 | GSPGTPGSRSRTPSL | |
| Tau iso8 | P10636-8 | S210 | PGTPGSRSRTPSLPT | |
| TTK | P33981 | S363 | LKNKTESSLLAKLEE | |
| TTK | P33981 | S37 | EDLTDELSLNKISAD | |
| TTK | P33981 | S436 | VFSVSKQSPPISTSK | |
| TTK | P33981 | S582 | LNKLQQHSDKIIRLY | + |
| TTK | P33981 | S677 | QMQPDTTSVVKDSQV | + |
| TTK | P33981 | S742 | QQIINQISKLHAIID | + |
| TTK | P33981 | S80 | EKNSVPLSDALLNKL | |
| TTK | P33981 | S821 | GQLVGLNSPNSILKA | ? |
| TTK | P33981 | S824 | VGLNSPNSILKAAKT | |
| TTK | P33981 | S845 | GGESHNSSSSKTFEK | |
| TTK | P33981 | S847 | ESHNSSSSKTFEKKR | |
| TTK | P33981 | T288 | SPDCDVKTDDSVVPC | ? |
| TTK | P33981 | T33 | KFKNEDLTDELSLNK | |
| TTK | P33981 | T360 | SITLKNKTESSLLAK | |
| TTK | P33981 | T564 | LEEADNQTLDSYRNE | |
| TTK | P33981 | T675 | ANQMQPDTTSVVKDS | + |
| TTK | P33981 | T676 | NQMQPDTTSVVKDSQ | + |
| TTK | P33981 | T686 | VKDSQVGTVNYMPPE | + |
| TTK | P33981 | T795 | HPYVQIQTHPVNQMA | |
| TTK | P33981 | T805 | VNQMAKGTTEEMKYV | |
| TTK | P33981 | T806 | NQMAKGTTEEMKYVL | - |
| TTK | P33981 | T849 | HNSSSSKTFEKKRGK | |
| TTK | P33981 | Y811 | GTTEEMKYVLGQLVG | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
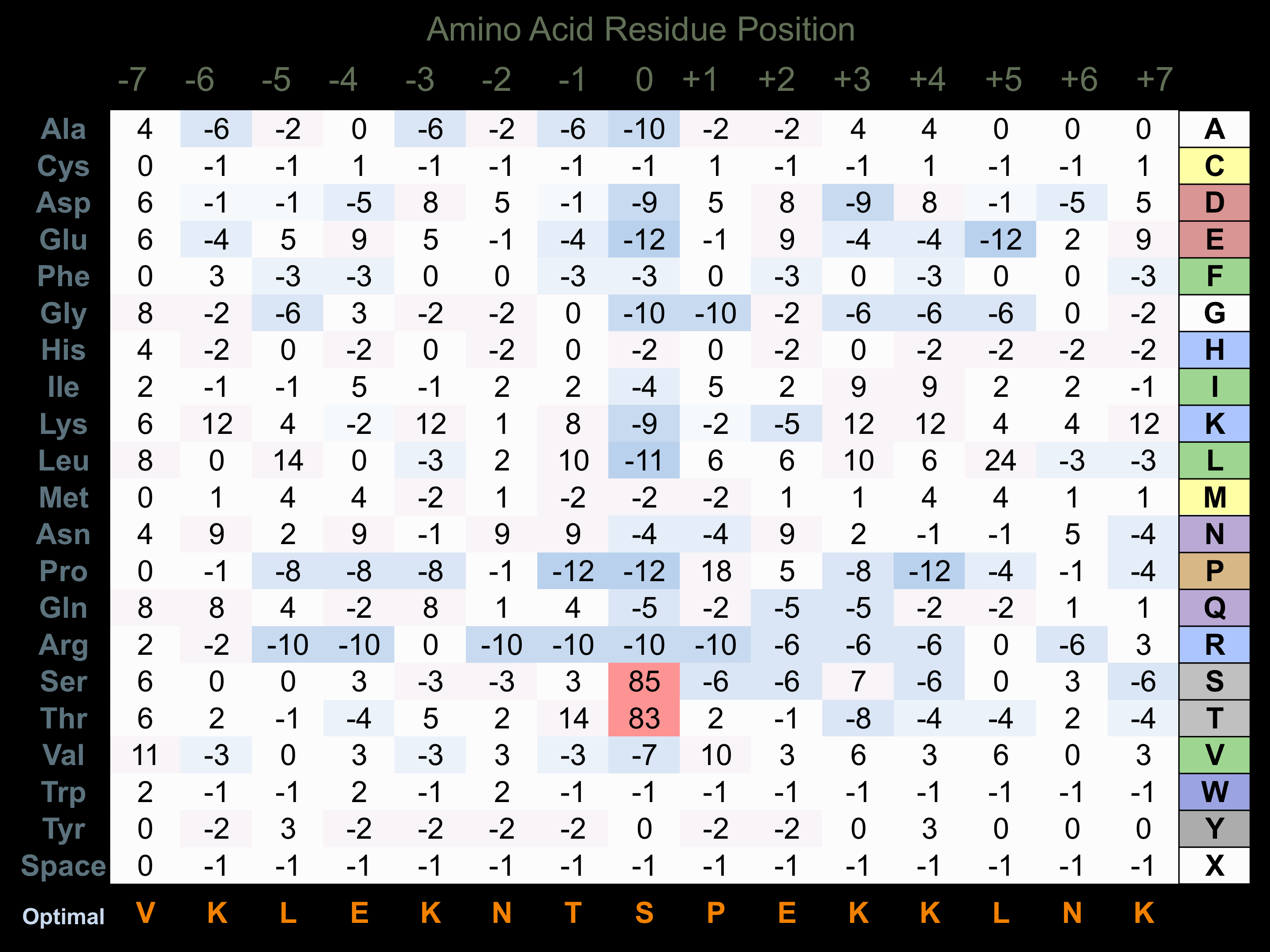
Matrix Type:
Experimentally derived from alignment of 42 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Pancreatic ductal adenocarcinomas (PDAC); Triple negative breast cancer (TNBC); Gastric cancer
Comments:
TTK is linked to pancreatic ductal adenocarcinoma (PDAC), which is a pancreatic cancer with high mortality rates. TTK is important in the growth and proliferation of PDAC cancer cells. Triple Negative Breast Cancer (TNBC) is a highly aggressive form of cancer and TTK is consistently upregulated in TNBC tissues.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +235, p<0.019); Bladder carcinomas (%CFC= +109, p<0.058); Brain glioblastomas (%CFC= -48, p<0.002); Breast non-basal-like cancer (BLC) (%CFC= +109, p<0.0001); Breast sporadic basal-like cancer (BLC) (%CFC= +167, p<0.0001); Cervical epithelial cancer (%CFC= -50, p<0.025); Colon mucosal cell adenomas (%CFC= +151, p<0.0001); Gastric cancer (%CFC= +85, p<0.0004); Large B-cell lymphomas (%CFC= +46, p<0.007); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +83, p<0.055); Oral squamous cell carcinomas (OSCC) (%CFC= +141, p<0.033); Ovary adenocarcinomas (%CFC= +828, p<0.002); and Vulvar intraepithelial neoplasia (%CFC= +424, p<0.0001). The COSMIC website notes an up-regulated expression score for TTK in diverse human cancers of 543, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 7 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. The phosphotransferase activity of TTK can be lost with a D664A or T686A mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 25316 diverse cancer specimens. This rate is only 31 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.9 % in 1093 large intestine cancers tested; 0.47 % in 589 stomach cancers tested; 0.33 % in 602 endometrium cancers tested; 0.19 % in 1755 lung cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,528 cancer specimens
Comments:
Over 60 deletions (37 at R854fs*>4 and 12 at K857fs*? located at the C-terminus), 2 insertions, and no complex mutations are noted on the COSMIC website.

