Nomenclature
Short Name:
TRAD
Full Name:
Serine-threonine-protein kinase Duet
Alias:
- Duet
- Duo
- Kalirin, RhoGEF kinase
- Serine/threonine kinase with Dbl- and pleckstrin homology domains
- Serine/threonine kinase with Dbl and pleckstriny domains
- KALRN
- EC 2.7.11.1
- HAPIP
- Hs.8004
- Huntingtin-associated protein-interacting protein
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
Trio
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
340,174
# Amino Acids:
2985
# mRNA Isoforms:
6
mRNA Isoforms:
340,174 Da (2985 AA; O60229); 192,229 Da (1663 AA; O60229-2); 144,485 Da (1289 AA; O60229-4); 141,164 Da (1257 AA; O60229-6); 98,659 Da (851 AA; O60229-3); 82,806 Da (738 AA; O60229-5)
4D Structure:
Interacts with the C-terminal of peptidylglycine alpha-amidating monooxygenase (PAM) and with the huntingtin-associated protein 1 (HAP1)
1D Structure:
Subfamily Alignment
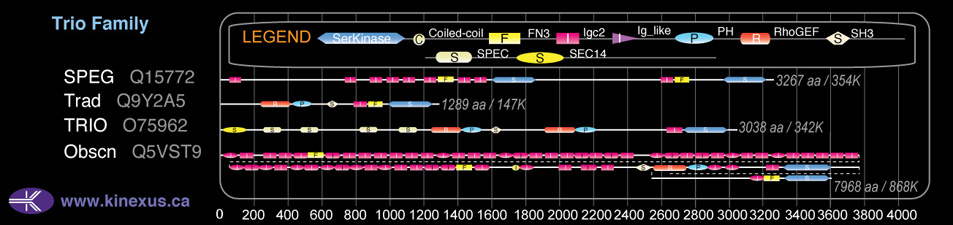
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 35 | 180 | CRAL-TRIO |
| 188 | 308 | Spectrin 1 |
| 310 | 416 | Spectrin 2 |
| 536 | 642 | Spectrin 3 |
| 890 | 1004 | Spectrin 4 |
| 1130 | 1222 | Spectrin 5 |
| 1281 | 1450 | RhoGEF |
| 1468 | 1580 | PH |
| 1646 | 1711 | SH3 |
| 1928 | 2103 | RhoGEF |
| 2115 | 2225 | PH |
| 2320 | 2385 | SH3 |
| 2470 | 2563 | Ig-like C2-type |
| 2568 | 2659 | Fibronectin type-III |
| 2683 | 2937 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K1182, K1187.
Methylated:
R1036.
Serine phosphorylated:
S86, S103, S223, S487, S672, S1256, S1607, S1750, S1753, S1756, S1763, S1773, S1780, S1799, S1817, S2236, S2261, S2436, S2559, S2715.
Threonine phosphorylated:
T1599+, T1912.
Tyrosine phosphorylated:
Y1079, Y1943, Y2240, Y2301.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1297
56
1351
 3
3
43
20
78
 17
17
222
58
555
 42
42
542
257
1232
 36
36
470
63
352
 1.3
1.3
17
110
21
 8
8
108
78
212
 57
57
735
105
1714
 19
19
243
20
238
 6
6
79
248
204
 11
11
142
90
520
 35
35
451
265
503
 10
10
135
80
315
 2
2
23
12
31
 13
13
171
84
608
 3
3
33
35
49
 3
3
38
558
139
 15
15
190
69
677
 17
17
221
214
368
 30
30
386
224
344
 12
12
151
83
331
 14
14
182
86
346
 23
23
292
60
478
 12
12
153
71
232
 10
10
136
83
420
 71
71
927
175
2617
 8
8
110
86
310
 11
11
149
68
460
 20
20
258
70
497
 16
16
213
84
245
 46
46
601
48
440
 47
47
614
62
1200
 8
8
104
167
426
 59
59
770
156
683
 5
5
70
96
67
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.4
98.4
98.8
0 -
-
-
98 -
-
-
98.5 -
-
-
- 97.4
97.4
98.3
99 -
-
-
- 97
97
98.3
98 97
97
98.3
98 -
-
-
- 61.7
61.7
76.4
- 91.8
91.8
95.2
94 -
-
-
75 60.9
60.9
75.3
- -
-
-
- 34
34
50.8
- -
-
-
- -
-
-
- 21.2
21.2
39.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by phosphorylation at Thr-95 and Thr-1599.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
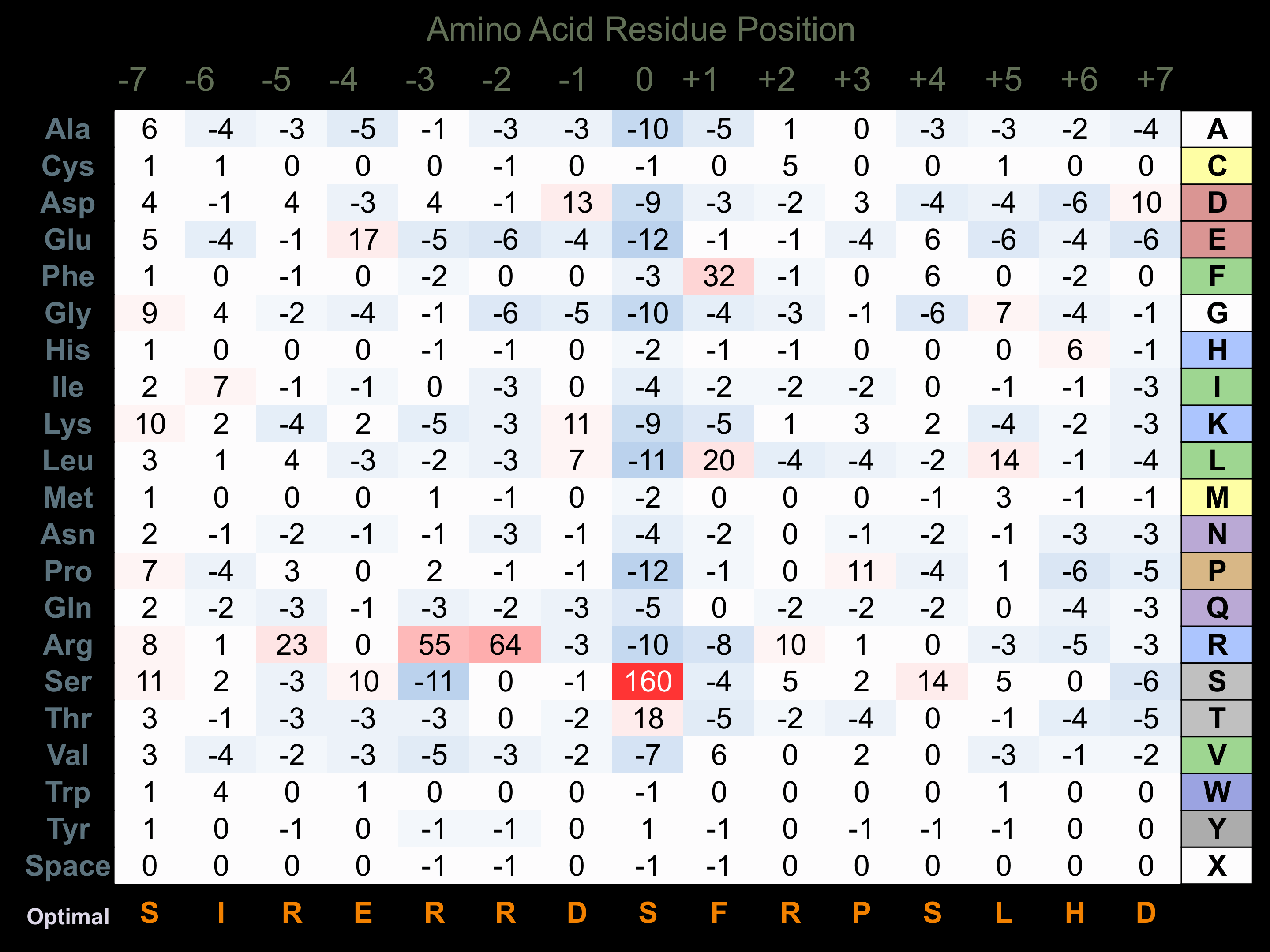
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Heart disorder
Specific Diseases (Non-cancerous):
Coronary heart disease 5 (CHDS5)
Comments:
Coronary Heart Disease 5 (CHDS5) is the result of lack of sufficient blood flow to the heart, leading to complications.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -52, p<0.0009); and Ovary adenocarcinomas (%CFC= +122, p<0.0006).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A K2712A mutation in TRAD can result in inhibited autophosphorylation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24434 diverse cancer specimens. This rate is very similar (+ 3% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.43 % in 864 skin cancers tested; 0.26 % in 589 stomach cancers tested; 0.25 % in 1229 large intestine cancers tested; 0.19 % in 603 endometrium cancers tested; 0.17 % in 548 urinary tract cancers tested; 0.17 % in 1609 lung cancers tested; 0.09 % in 273 cervix cancers tested; 0.09 % in 1512 liver cancers tested; 0.08 % in 710 oesophagus cancers tested; 0.07 % in 238 bone cancers tested; 0.05 % in 942 upper aerodigestive tract cancers tested; 0.04 % in 881 prostate cancers tested; 0.04 % in 1289 breast cancers tested; 0.03 % in 1982 haematopoietic and lymphoid cancers tested; 0.03 % in 1437 pancreas cancers tested; 0.03 % in 1253 kidney cancers tested; 0.02 % in 807 ovary cancers tested; 0.02 % in 558 thyroid cancers tested; 0.02 % in 441 autonomic ganglia cancers tested; 0.01 % in 382 soft tissue cancers tested; 0.01 % in 2030 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R313W (4); R520W (4); R520Q (2).
Comments:
Only 8 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website. About 39% of the point mutations are silent and do not change the amino acid sequence of the protein kinase.

