Nomenclature
Short Name:
MYLK2
Full Name:
Myosin light chain kinase 2, skeletal-cardiac muscle
Alias:
- EC 2.7.11.18
- KMLC
- SkMLCK
- MLCK2
- Myosin light chain kinase 2
- Myosin light chain kinase 2, skeletal/cardiac muscle
- Skeletal muscle myosin light chain kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
MLCK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
64685
# Amino Acids:
596
# mRNA Isoforms:
1
mRNA Isoforms:
64,685 Da (596 AA; Q9H1R3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
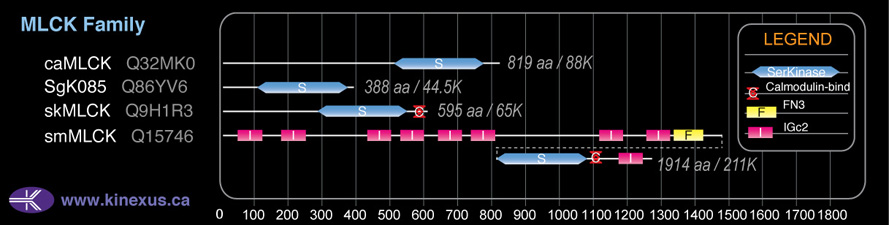
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 285 | 540 | Pkinase |
| 573 | 585 | CaM_binding |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Ubiquitinated:
K584.
Acetylated:
A2.
Serine phosphorylated:
S143, S150, S224, S586, S587, S588.
Tyrosine phosphorylated:
Y563.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 38
38
1286
9
2290
 0.1
0.1
4
4
3
 8
8
258
11
583
 0.8
0.8
27
39
44
 9
9
313
13
283
 0.2
0.2
8
9
6
 0.1
0.1
3
13
1
 2
2
82
16
136
 0.1
0.1
2
3
0
 1.5
1.5
49
37
87
 2
2
67
16
92
 14
14
485
22
671
 2
2
75
11
94
 0.1
0.1
2
3
1
 3
3
107
5
153
 0.2
0.2
7
5
7
 3
3
111
79
88
 5
5
164
14
360
 40
40
1336
46
891
 11
11
370
31
252
 2
2
68
16
97
 2
2
68
16
106
 4
4
118
11
233
 1.4
1.4
48
14
70
 2
2
83
16
125
 15
15
487
30
630
 3
3
101
14
187
 2
2
68
14
112
 4
4
145
14
416
 26
26
863
14
196
 29
29
979
12
34
 100
100
3358
11
3658
 0.5
0.5
16
12
3
 27
27
922
26
800
 4
4
120
22
106
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 37.3
37.3
49.5
99 37.9
37.9
50.9
95 -
-
-
85 -
-
-
- 81.7
81.7
86.1
87 -
-
-
- 80.9
80.9
85.2
84 80.8
80.8
85.4
82 -
-
-
- 42
42
54.7
- 48.5
48.5
58.3
68 -
-
-
- 40.1
40.1
56.2
- -
-
-
- -
-
-
52 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| MRLC1 (MYL9) | P24844 | S20 | KRPQRATSNVFAMFD | |
| MRLC1 (MYL9) | P24844 | T19 | KKRPQRATSNVFAMF | |
| MYL7 | Q01449 | S22 | TKQAQRGSSNVFSMF | |
| MYL7 | Q01449 | S23 | KQAQRGSSNVFSMFE | |
| MYLK2 (skMLCK) | Q9H1R3 | S150 | PAFLHSPSCPAIISS | |
| MYLK2 (skMLCK) | Q9H1R3 | S224 | AKMQGDTSRGIEFQA |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
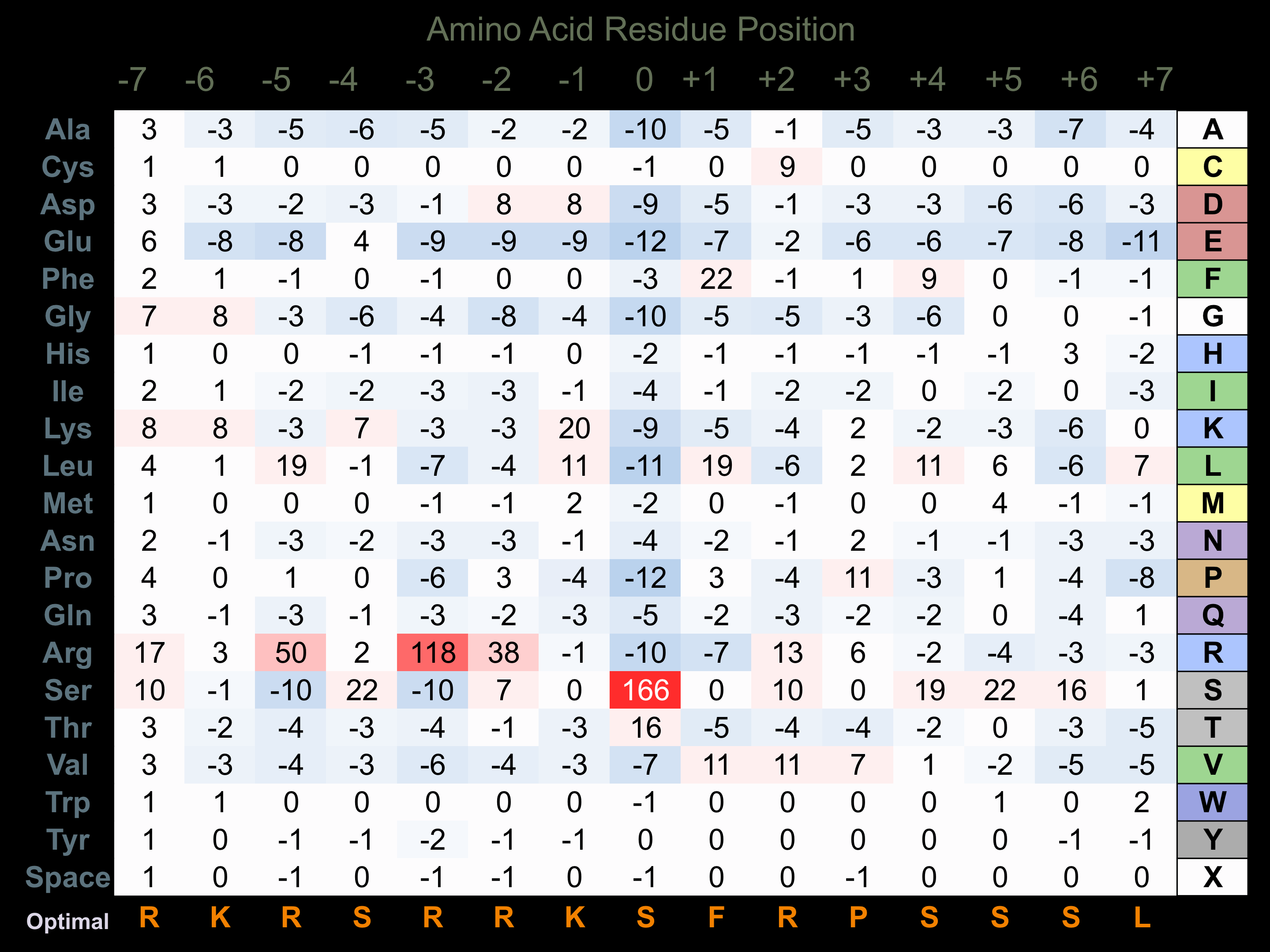
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cardiovascular disorders
Specific Diseases (Non-cancerous):
Hypertrophic cardiomyopathy, midventricular, digenic; Familial hypertrophic cardiomyopathy; Caveolinopathies; Cardiomyopathy, familial hypertrophic, 1
Comments:
Familial hypertrophic cardiomyopathy is an hereditary cardiovascular disease characterized by the hypertrophy (thickening) of the heart muscle, specifically in the left ventricular wall. The disease primarily affects the myocardium and results in the asymmetric thickening of this muscular layer leading to dysfunction of the heart and potential sudden death without prior symptoms. Myocardial thickening is usually most evident in the interventricular septum, which can reduce or completely prevent the outflow of blood from the heart. The symptoms of the disease include dyspnea, syncope, collapse, heart palpitations, and chest pain, although many reported cases were asymptomatic and characterized by a sudden onset. Heart tissue analysis from affected individuals displays myocyte disarray and fibrosis. Caveolinopathies are a group of muscular disorders that can be separated into 5 phenotypic categories and characterized by muscle weakness, muscle cramps, elevated serum levels of creatine kinase (CK), and increased muscle irritability. In ~99% of patients with these disorders there are mutations in the gene that encodes for caveolin-3, a membrane protein specific to muscle cells. Several mutations in myosin RLK have been reported in patients with familial hypertrophic cardiomyopathy, affecting the structure, phosphorylation, and calcium binding properties of the proteins. Therefore, as an upstream regualtor of the myosin RLK protein, aberrant activity of MYLK2 has been linked to development of hypertrophic cardiomyopathy. In agreement with this, mutations in the MYLK2 gene have been observed in patients with hypertrophic cardiomyopathy. For example, a double point mutation was observed in a 13 year old white male with midventricular hypertrophic cardiomyopathy, including A87V and A95E substitution mutations. In vitro enzymatic analysis of the mutant MYLK2 protein revealed an approximately doubled Vmax compared to the wildtype protein, which was suggested to be responsible for stimulating the observed hypertrophy in the myocardial tissue.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Prostate cancer - metastatic (%CFC= +55, p<0.001). The COSMIC website notes an up-regulated expression score for MYLK2 in diverse human cancers of 399, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 1 for this protein kinase in human cancers was 98% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 24726 diverse cancer specimens. This rate is only 32 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.62 % in 864 skin cancers tested; 0.4 % in 1270 large intestine cancers tested; 0.25 % in 603 endometrium cancers tested; 0.21 % in 548 urinary tract cancers tested; 0.16 % in 1634 lung cancers tested; 0.14 % in 589 stomach cancers tested; 0.14 % in 1512 liver cancers tested; 0.13 % in 127 biliary tract cancers tested; 0.1 % in 833 ovary cancers tested; 0.08 % in 1316 breast cancers tested; 0.07 % in 710 oesophagus cancers tested; 0.05 % in 1459 pancreas cancers tested; 0.04 % in 942 upper aerodigestive tract cancers tested; 0.04 % in 881 prostate cancers tested; 0.04 % in 441 autonomic ganglia cancers tested; 0.03 % in 1276 kidney cancers tested; 0.02 % in 2082 central nervous system cancers tested; 0.02 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,009 cancer specimens
Comments:
Only 1 complex mutation, and no deletion or insertions are noted on the COSMC website.

