Nomenclature
Short Name:
MAPKAPK2
Full Name:
MAP kinase-activated protein kinase 2
Alias:
- EC 2.7.11.1
- MAP kinase-activated protein kinase 2
- MAPK2
- MAPKAP kinase 2
- MK2
- RPS6KC1
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
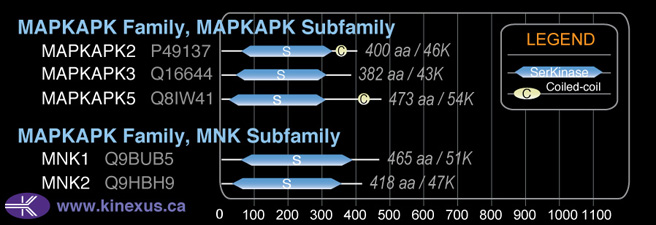
MAPKAPK
SubFamily:
MAPKAPK
Specific Links
Structure
Mol. Mass (Da):
45,568
# Amino Acids:
400
# mRNA Isoforms:
2
mRNA Isoforms:
45,568 Da (400 AA; P49137); 42,203 Da (370 AA; P49137-2)
4D Structure:
Interacts with PHC2.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 64 | 325 | Pkinase |
| 341 | 364 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S9, S67, S219, S272+, S328, S339.
Sumoylated:
K353.
Threonine phosphorylated:
T25, T66, T206, T214, T215, T221, T222+, T226-, T317, T329, T334+, T338.
Tyrosine phosphorylated:
Y63, Y225+, Y228-, Y229-, Y367.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 33
33
1322
39
1714
 4
4
178
26
160
 8
8
336
19
266
 7
7
290
143
437
 23
23
912
36
723
 2
2
81
112
87
 12
12
487
55
695
 21
21
857
69
719
 14
14
583
24
526
 7
7
287
159
326
 4
4
180
55
161
 17
17
672
305
617
 6
6
231
52
249
 4
4
157
24
147
 7
7
269
47
294
 4
4
178
25
138
 5
5
200
395
162
 7
7
287
35
279
 10
10
422
145
530
 14
14
578
162
591
 6
6
256
44
246
 9
9
358
50
373
 12
12
469
38
468
 9
9
353
36
319
 7
7
268
44
234
 16
16
637
88
627
 6
6
261
55
309
 7
7
273
36
255
 6
6
238
36
242
 10
10
398
42
259
 16
16
645
42
617
 100
100
4025
55
8002
 4
4
158
51
144
 19
19
776
88
684
 5
5
199
57
295
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.8
100 98.8
98.8
98.8
98 -
-
-
99 -
-
-
99 76.3
76.3
76.5
97 -
-
-
- 93.3
93.3
94.8
98 65.5
65.5
79.3
97.5 -
-
-
- 75.5
75.5
77
- 71.5
71.5
73.8
93 81.3
81.3
86
90 27.7
27.7
46.2
89 -
-
-
- 55.8
55.8
71.3
- 55.6
55.6
70.1
- -
-
-
50 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK14 - Q16539 |
| 2 | HSPB1 - P04792 |
| 3 | YWHAZ - P63104 |
| 4 | ZFP36 - P26651 |
| 5 | CDC25A - P30304 |
| 6 | LSP1 - P33241 |
| 7 | CREB1 - P16220 |
| 8 | TCF3 - P15923 |
| 9 | HNRNPA0 - Q13151 |
| 10 | TSC2 - P49815 |
| 11 | ETV1 - P50549 |
| 12 | EGF - P01133 |
| 13 | AKT1 - P31749 |
| 14 | SRF - P11831 |
| 15 | MAPK1 - P28482 |
Regulation
Activation:
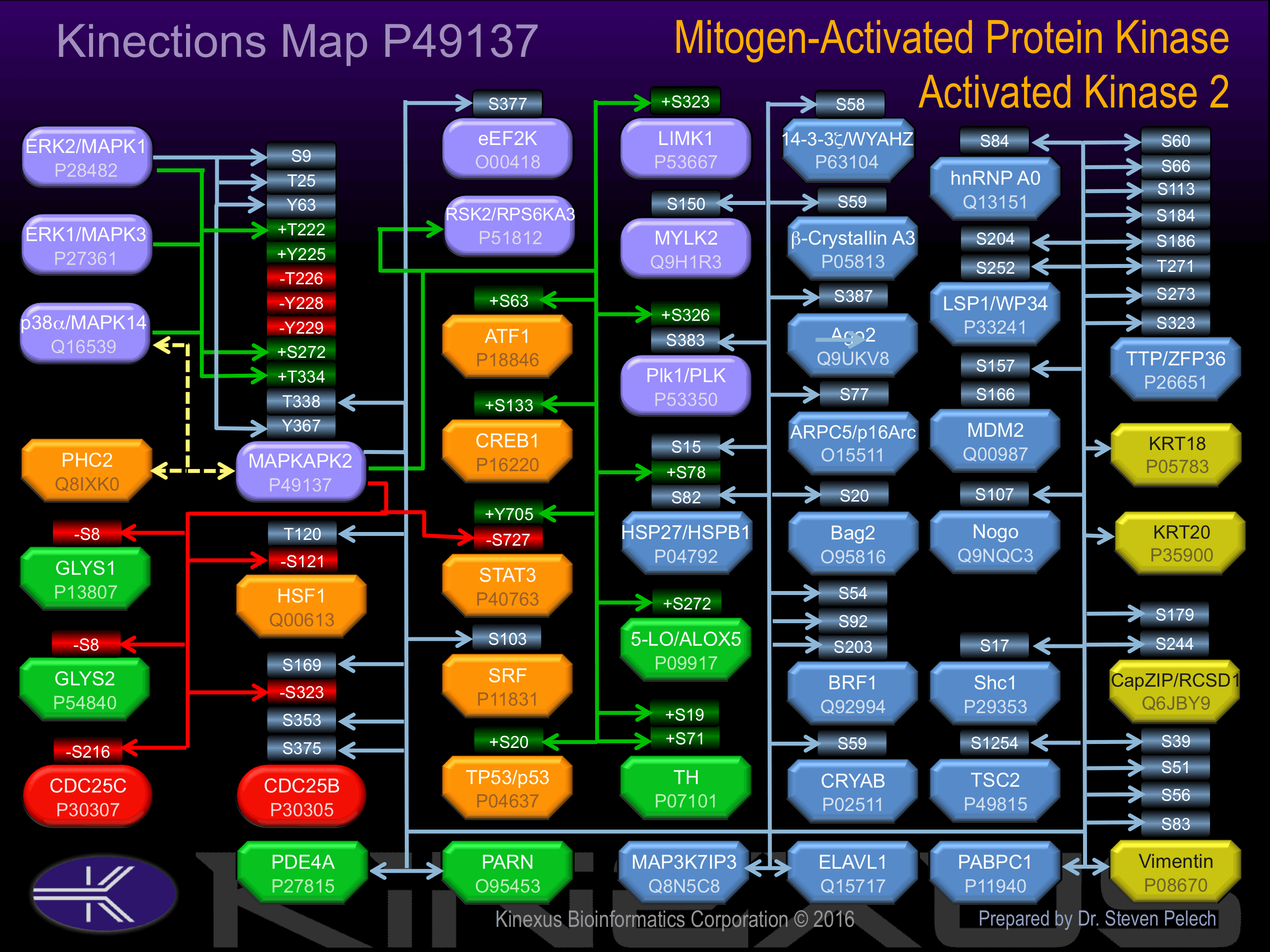
Phosphorylation of Thr-222, Thr-334 and Ser-272 increases phosphotransferase activity. Seems to be activated by two distinct pathways: the first involves the stimulation of p42/p44 MAPK by growth factors, the second, triggered by stress and heat shock, depends on the activation of MPK2 and upstream MAPKK/MAPKKK.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ERK2 | P28482 | S9 | LSNSQGQSPPVPFPA | |
| ERK2 | P28482 | T25 | APPPQPPTPALPHPP | |
| p38a | Q16539 | T25 | APPPQPPTPALPHPP | |
| ERK2 | P28482 | Y63 | KNAIIDDYKVTSQVL | |
| ERK2 | P28482 | T222 | TSHNSLTTPCYTPYY | + |
| p38a | Q16539 | T222 | TSHNSLTTPCYTPYY | + |
| ERK1 | P27361 | T222 | TSHNSLTTPCYTPYY | + |
| ERK2 | P28482 | S272 | SNHGLAISPGMKTRI | + |
| p38a | Q16539 | S272 | SNHGLAISPGMKTRI | + |
| ERK2 | P28482 | T334 | QSTKVPQTPLHTSRV | + |
| p38a | Q16539 | T334 | QSTKVPQTPLHTSRV | + |
| ERK1 | P27361 | T334 | QSTKVPQTPLHTSRV | + |
| MAPKAPK2 | P49137 | T338 | VPQTPLHTSRVLKED | |
| ERK2 | P28482 | Y367 | LATMRVDYEQIKIKK |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 14-3-3 zeta (YWHAZ) | P63104 | S58 | VVGARRSSWRVVSSI | |
| 5-LO (ALOX5) | P09917 | S272 | CSLERQLSLEQEVQQ | + |
| Ago2 | Q9UKV8 | S387 | SKLMRSASFNTDPYV | |
| ARPC5 (p16 Arc) | O15511 | S77 | AVKDRAGSIVLKVLI | |
| ATF1 | P18846 | S63 | GILARRPSYRKILKD | + |
| Bag2 | O95816 | S20 | GRFCRSSSMADRSSR | |
| Beta-crystallin A3 | P05813 | S59 | PSFLRAPSWIDTGLS | |
| BRF1 | Q92994 | S203 | PRLQHSFSFAGFPSA | |
| BRF1 | Q92994 | S54 | GGFPRRHSVTLPSSK | |
| BRF1 | Q92994 | S92 | RFRDRSFSEGGERLL | |
| CapZIP (RCSD1) | Q6JBY9 | S179 | RRFRRSQSDCGELGD | |
| CapZIP (RCSD1) | Q6JBY9 | S244 | PPLRRSPSRTEKQEE | |
| Cdc25B | P30305 | S169 | VLRNITNSQAPDGRR | |
| Cdc25B | P30305 | S323 | QRLFRSPSMPCSVIR | - |
| Cdc25B | P30305 | S353 | VQNKRRRSVTPPEEQ | |
| Cdc25B | P30305 | S375 | ARVLRSKSLCHDEIE | |
| Cdc25C | P30307 | S216 | SGLYRSPSMPENLNR | ? |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| CRYAB | P02511 | S59 | PSFLRAPSWFDTGLS | ? |
| eEF2K | O00418 | S377 | PPLLRPLSENSGDEN | |
| GYS1 | P13807 | S8 | MPLNRTLSMSSLPGL | - |
| GYS2 | P54840 | S8 | MLRGRSLSVTSLGGL | - |
| hnRNP A0 | Q13151 | S84 | VELKRAVSREDSARP | |
| HSF1 | Q00613 | S121 | NIKRKVTSVSTLKSE | - |
| HSF1 | Q00613 | T120 | ENIKRKVTSVSTLKS | |
| HSP27 (HSPB1) | P04792 | S15 | FSLLRGPSWDPFRDW | ? |
| HSP27 (HSPB1) | P04792 | S78 | PAYSRALSRQLSSGV | + |
| HSP27 (HSPB1) | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| LIMK1 | P53667 | S323 | KDLGRSESLRVVCRP | + |
| LSP1 (WP34) | P33241 | S204 | KILDRTESLNRSIEK | |
| LSP1 (WP34) | P33241 | S252 | PKLARQASIELPSMA | |
| MAPKAPK2 | P49137 | T338 | VPQTPLHTSRVLKED | |
| MDM2 | Q00987 | S157 | SHLVSRPSTSSRRRA | |
| MDM2 | Q00987 | S166 | SSRRRAISETEENSD | |
| MYLK2 (skMLCK) | Q9H1R3 | S150 | PAFLHSPSCPAIISS | |
| Nogo | Q9NQC3 | S107 | VAPERQPSWDPSPVS | |
| p53 | P04637 | S20 | PLSQETFSDLWKLLP | + |
| Plk1 (PLK) | P53350 | S326 | LTIPPRFSIAPSSLD | + |
| Plk1 (PLK) | P53350 | S383 | DMLQQLHSVNASKPS | |
| Shc1 | P29353 | S17 | YNPLRNESLSSLEEG | |
| SRF | P11831 | S103 | RGLKRSLSEMEIGMV | |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| STAT3 | P40763 | Y705 | DPGSAAPYLKTKFIC | + |
| TH | P07101 | S19 | KGFRRAVSEQDAKQA | + |
| TH | P07101 | S71 | RFIGRRQSLIEDARK | + |
| TSC2 | P49815 | S1254 | TALYKSLSVPAASTA | |
| TTP (ZFP36) | P26651 | S113 | TELCRTFSESGRCRY | |
| TTP (ZFP36) | P26651 | S184 | HPPVLRQSISFSGLP | |
| TTP (ZFP36) | P26651 | S186 | PVLRQSISFSGLPSG | |
| TTP (ZFP36) | P26651 | S273 | GGLVRTPSVQSLGSD | |
| TTP (ZFP36) | P26651 | S323 | LPIFNRISVSE____ | |
| TTP (ZFP36) | P26651 | S60 | RLPGRSTSLVEGRSC | |
| TTP (ZFP36) | P26651 | S66 | TSLVEGRSCGWVPPP | |
| TTP (ZFP36) | P26651 | T271 | PLGGLVRTPSVQSLG | |
| Vimentin | P08670 | S39 | TTSTRTYSLGSALRP | |
| Vimentin | P08670 | S51 | LRPSTSRSLYASSPG | |
| Vimentin | P08670 | S56 | SRSLYASSPGGVYAT | |
| Vimentin | P08670 | S83 | GVRLLQDSVDFSLAD |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
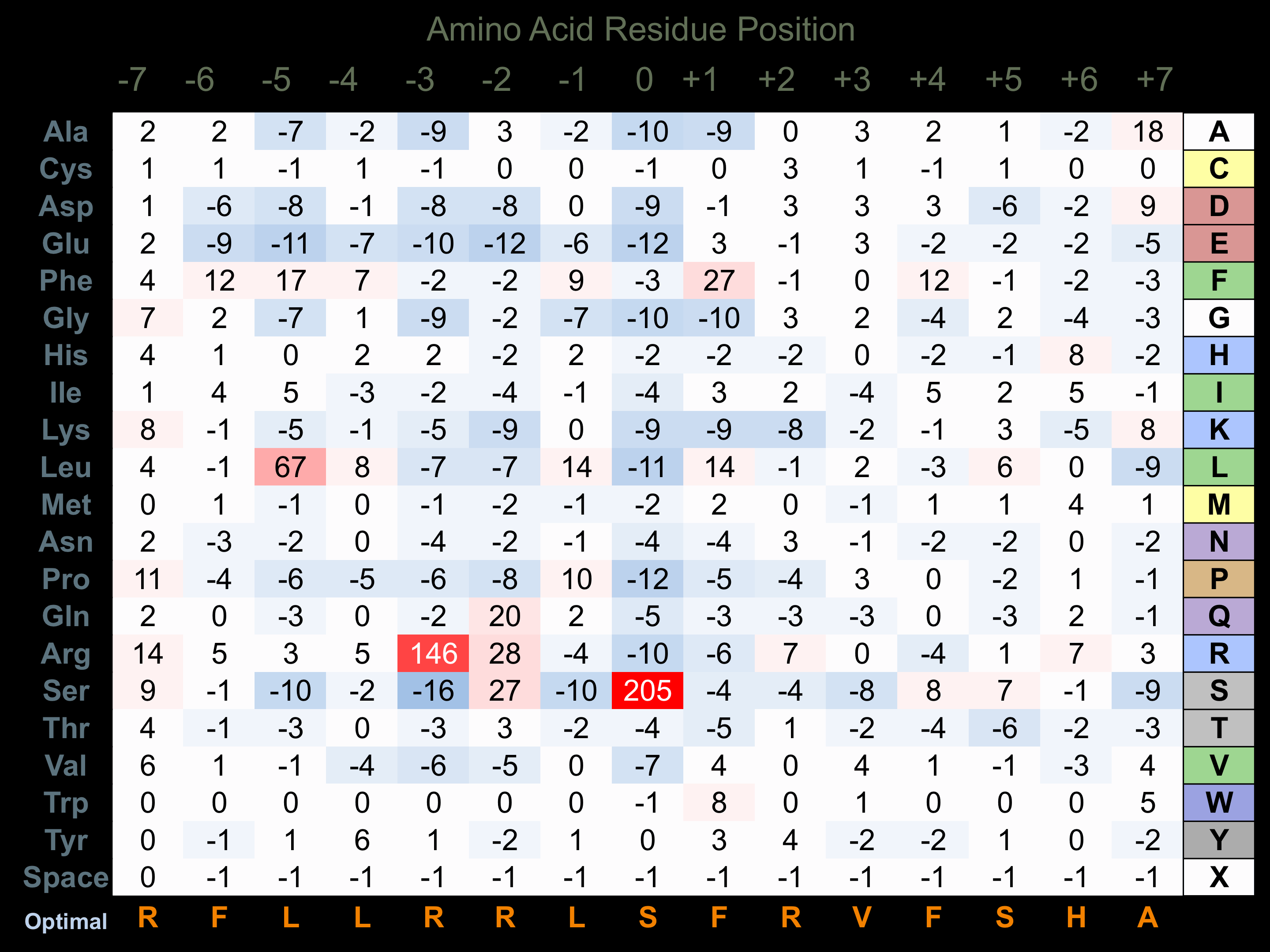
Matrix Type:
Experimentally derived from alignment of 73 known protein substrate phosphosites and 15 peptides phosphorylated by recombinant MAPKAPK2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Lung cancer (LC)
Comments:
MAPKAPK2 may be a tumour requiring protein (TRP). MAPKAPK2 has also been implicated in the promotion of cancer development and tumorigenesis, specifically lung cancer. The copy number variant g. CNV-30450 found in the sequence of the MAPKAP2 gene promoter has been associated with lung cancer. For example, individuals with 4 copies of the variant were found to have a significantly elevated risk of lung cancer compared to individuals with 2 or 3 copies of the variant. The mean survival time for lung cancer patients with 4 copies of the variant was also significantly lower (
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer stage 1B (%CFC= +163); Cervical cancer stage 2A (%CFC= +308); Cervical cancer stage 2B (%CFC= +157); Ovary adenocarcinomas (%CFC= +101, p<0.024); Skin melanomas - malignant (%CFC= +71, p<0.015); and Uterine fibroids (%CFC= +75, p<0.026). The COSMIC website notes an up-regulated expression score for MAPKAPK2 in diverse human cancers of 766, which is 1.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 77 for this protein kinase in human cancers was 1.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25183 diverse cancer specimens. This rate is only -9 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.34 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,466 cancer specimens
Comments:
Only 1 deletion and 1 insertion and no complex mutations are noted on the COSMIC website.