Nomenclature
Short Name:
BLK
Full Name:
Tyrosine-protein kinase BLK
Alias:
- EC 2.7.10.2
- P55-BLK
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Src
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
57,706
# Amino Acids:
505
# mRNA Isoforms:
1
mRNA Isoforms:
57,706 Da (505 AA; P51451)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
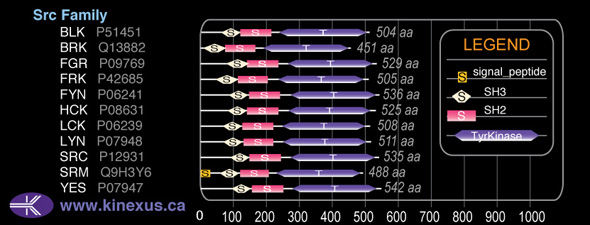
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Myristoylated:
G2 (predicted).
Serine phosphorylated:
S197, S341, S356.
Tyrosine phosphorylated:
Y107, Y187, Y188, Y205, Y309, Y314, Y350, Y389+, Y494, Y501-.
Ubiquitinated:
K157, K208, K305, K325, K375.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 65
65
594
29
1134
 2
2
20
16
29
 8
8
74
14
187
 37
37
336
93
886
 24
24
224
27
210
 6
6
58
81
183
 16
16
146
34
374
 100
100
918
45
2062
 20
20
186
20
169
 5
5
48
73
75
 4
4
34
34
67
 51
51
468
165
487
 19
19
170
36
234
 1.4
1.4
13
14
19
 3
3
30
28
47
 1.1
1.1
10
14
12
 4
4
37
122
112
 5
5
43
24
94
 5
5
43
92
57
 24
24
217
112
260
 6
6
51
27
120
 17
17
157
32
147
 8
8
72
24
170
 4
4
35
25
87
 7
7
66
26
143
 84
84
769
61
1589
 13
13
115
42
119
 7
7
65
24
151
 5
5
42
25
121
 12
12
111
28
106
 56
56
514
24
540
 95
95
875
42
911
 16
16
146
67
409
 62
62
570
57
545
 5
5
49
35
45
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
99 94.3
94.3
95.6
- -
-
-
87 -
-
-
94 87.9
87.9
94.5
89 -
-
-
- 85.9
85.9
92.7
87 61.1
61.1
76.3
88 -
-
-
- 61.5
61.5
77
- 64
64
78.5
73.5 54.8
54.8
69.1
67 65
65
78.2
69 -
-
-
- 51.6
51.6
66.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CBL - P22681 |
| 2 | CD79A - P11912 |
| 3 | UBE3A - Q05086 |
| 4 | CD79B - P40259 |
| 5 | BCAS2 - O75934 |
| 6 | FCGR2B - P31994 |
| 7 | BCR - P11274 |
| 8 | FCGR2A - P12318 |
| 9 | PLCG2 - P16885 |
| 10 | BCL2L1 - Q07817 |
| 11 | BCL2 - P10415 |
| 12 | ERBB2 - P04626 |
| 13 | EGFR - P00533 |
Regulation
Activation:
NA
Inhibition:
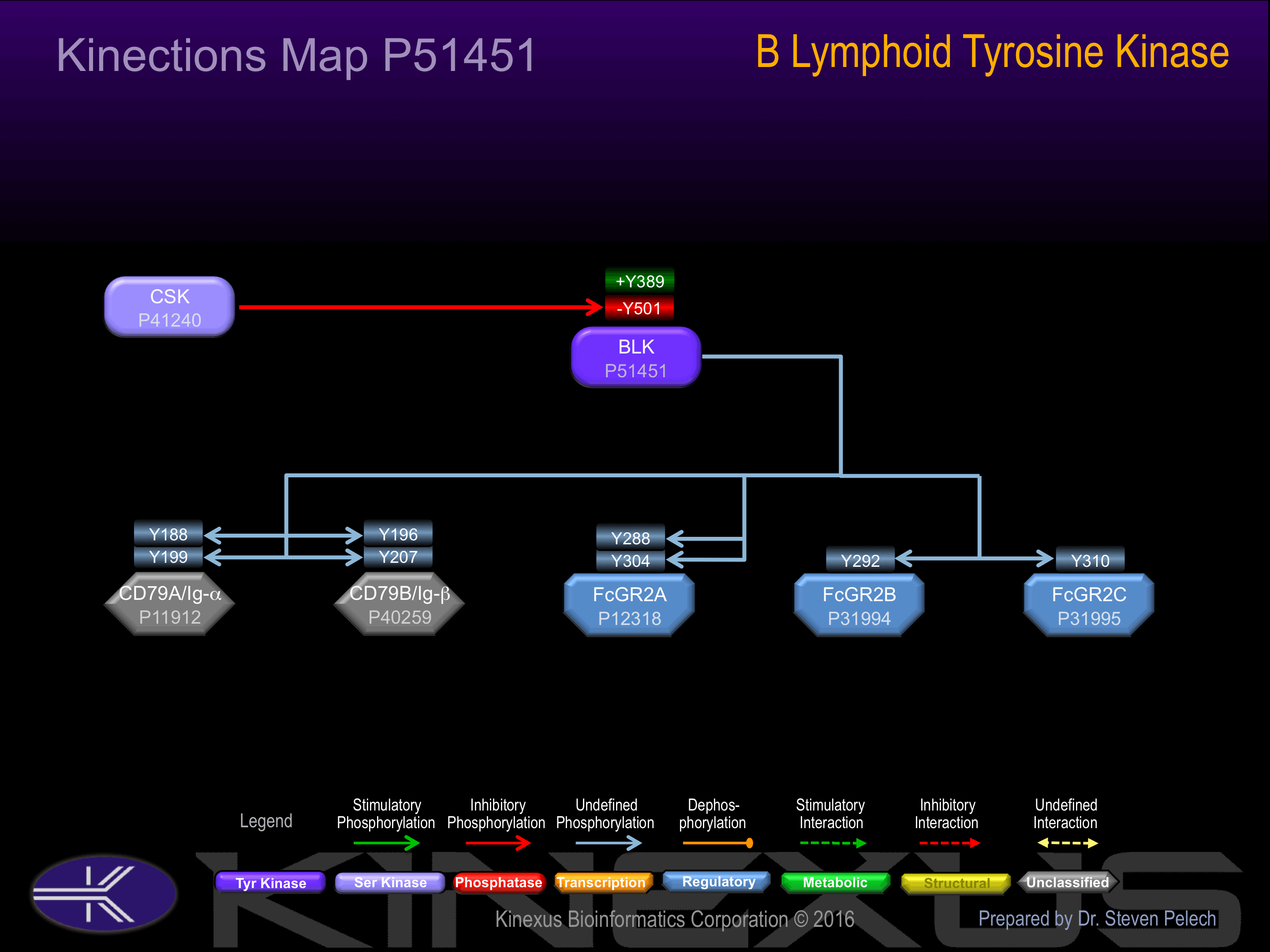
Phosphorylation of Tyr-501 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CD79A (Ig-alpha) | P11912 | Y188 | EYEDENLYEGLNLDD | |
| CD79A (Ig-alpha) | P11912 | Y199 | NLDDCSMYEDISRGL | |
| CD79B (Ig-beta) | P40259 | Y196 | GMEEDHTYEGLDIDQ | |
| CD79B (Ig-beta) | P40259 | Y207 | DIDQTATYEDIVTLR | |
| FcGR2A | P12318 | Y288 | YETADGGYMTLNPRA | |
| FcGR2A | P12318 | Y304 | TDDDKNIYLTLPPND | |
| FcGR2B | P31994 | Y292 | GAENTITYSLLMHPD | |
| FcGR2C | P31995 | Y310 | TDDDKNIYLTLPPND |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
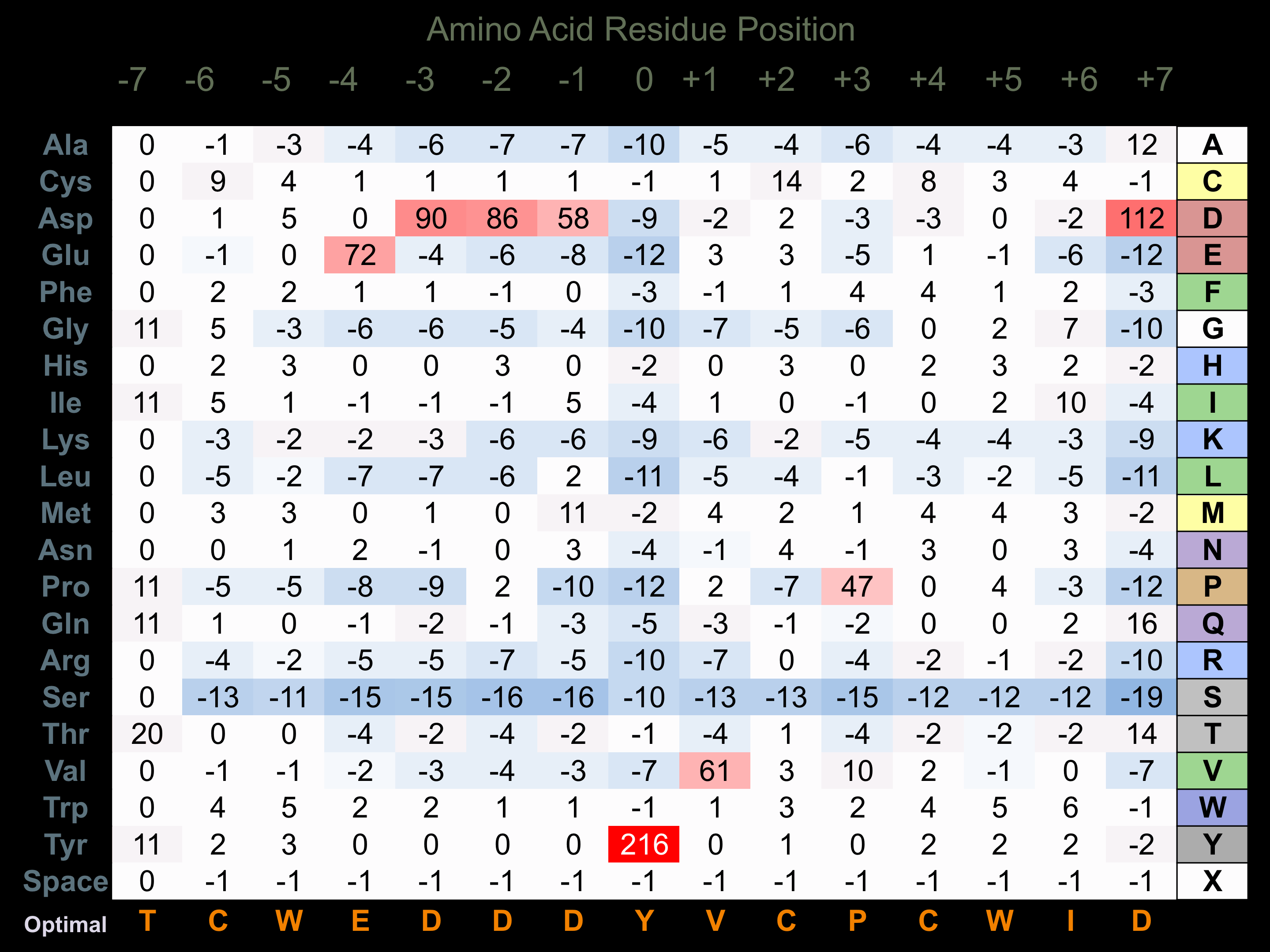
Matrix Type:
Experimentally derived from alignment of 7 known protein substrate phosphosites and 88 peptides phosphorylated by recombinant Blk in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological, diabetes and immune disorders
Specific Diseases (Non-cancerous):
Multifocal motor neuropathy; Sle susceptibility; Keratolytic winter erythema; Maturity-onset diabetes of the young, Type 1; Maturity-onset diabetes of the young, Type 11 (MODY11)
Comments:
An A71T mutation is associated with maturity-onset diabetes of Young 11 (MODY11), and results in reducing the enhancing effects of BLK on the expression of PDX1 and NKX6-1 and on insulin secretion. MODY11 is an early onset (usually before 25 years of age) form of diabetes that is autosomal dominant, and leads to a primary defect in insulin secretion.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Classical Hodgkin lymphomas (%CFC= -82, p<0.027); and Gastric cancer (%CFC= -62, p<0.0008). The COSMIC website notes an up-regulated expression score for BLK in diverse human cancers of 212, which is 0.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 11 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 25101 diverse cancer specimens. This rate is a modest 1.4-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.78 % in 864 skin cancers tested; 0.37 % in 589 stomach cancers tested; 0.36 % in 1270 large intestine cancers tested; 0.26 % in 603 endometrium cancers tested; 0.2 % in 1822 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,384 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.