Nomenclature
Short Name:
CSK
Full Name:
Tyrosine-protein kinase CSK
Alias:
- CYL
- EC 2.7.10.2
- MGC117393
- MPK-2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Csk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
50,704
# Amino Acids:
450
# mRNA Isoforms:
1
mRNA Isoforms:
50,704 Da (450 AA; P41240)
4D Structure:
Interacts with PTPN8 By similarity. Interacts with phosphorylated SIT1, PAG1, LIME1 and TGFB1I1. Interacts with SRCIN1
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
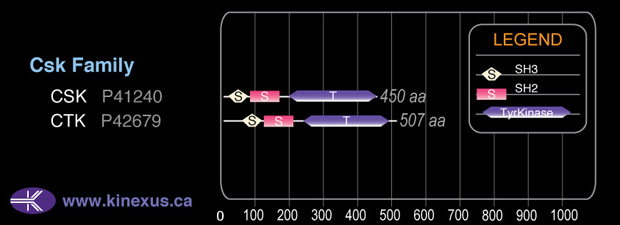
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
S2.
Serine phosphorylated:
S186, S284, S364+.
Threonine phosphorylated:
T23.
Tyrosine phosphorylated:
Y64, Y97, Y184+, Y263+, Y277+, Y304+, Y416.
Ubiquitinated:
K43, K171, K193, K196, K203, K222, K329, K337, K347, K393.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 42
42
940
16
1130
 6
6
141
10
131
 4
4
84
13
72
 57
57
1273
66
2212
 37
37
831
14
772
 6
6
142
37
55
 23
23
501
19
704
 76
76
1697
37
4064
 25
25
544
10
472
 10
10
214
45
184
 9
9
194
26
197
 32
32
704
117
659
 15
15
344
24
129
 5
5
111
9
104
 5
5
120
23
132
 9
9
193
8
101
 11
11
234
114
119
 6
6
144
20
147
 4
4
94
62
119
 31
31
686
56
645
 8
8
173
22
175
 27
27
595
24
734
 9
9
210
22
172
 4
4
94
20
122
 14
14
302
22
288
 100
100
2220
47
3077
 16
16
363
27
269
 7
7
149
20
131
 5
5
105
20
96
 3
3
69
14
56
 72
72
1592
18
672
 67
67
1478
21
1541
 9
9
203
56
315
 50
50
1109
31
793
 3
3
76
22
63
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.4
98.4
98.4
98 99.8
99.8
100
100 -
-
-
99 -
-
-
98 95.5
95.5
96.1
99 -
-
-
- 98.4
98.4
99.3
99 98.4
98.4
99.8
98 -
-
-
- -
-
-
- 93.3
93.3
95.3
93 83.8
83.8
90.4
84 85.8
85.8
93.1
86 -
-
-
- 26.6
26.6
32.8
63.5 48.3
48.3
62.9
- 43.4
43.4
57.3
53 36.8
36.8
45.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | SRC - P12931 |
| 2 | FYN - P06241 |
| 3 | SHC1 - P29353 |
| 4 | PXN - P49023 |
| 5 | RGS16 - O15492 |
| 6 | DOK1 - Q99704 |
| 7 | ADRB2 - P07550 |
| 8 | PTPN12 - Q05209 |
| 9 | FGR - P09769 |
| 10 | HNRNPK - P61978 |
| 11 | HCK - P08631 |
| 12 | CD44 - P16070 |
| 13 | GSN - P06396 |
| 14 | PTPN18 - Q99952 |
| 15 | LCK - P06239 |
Regulation
Activation:
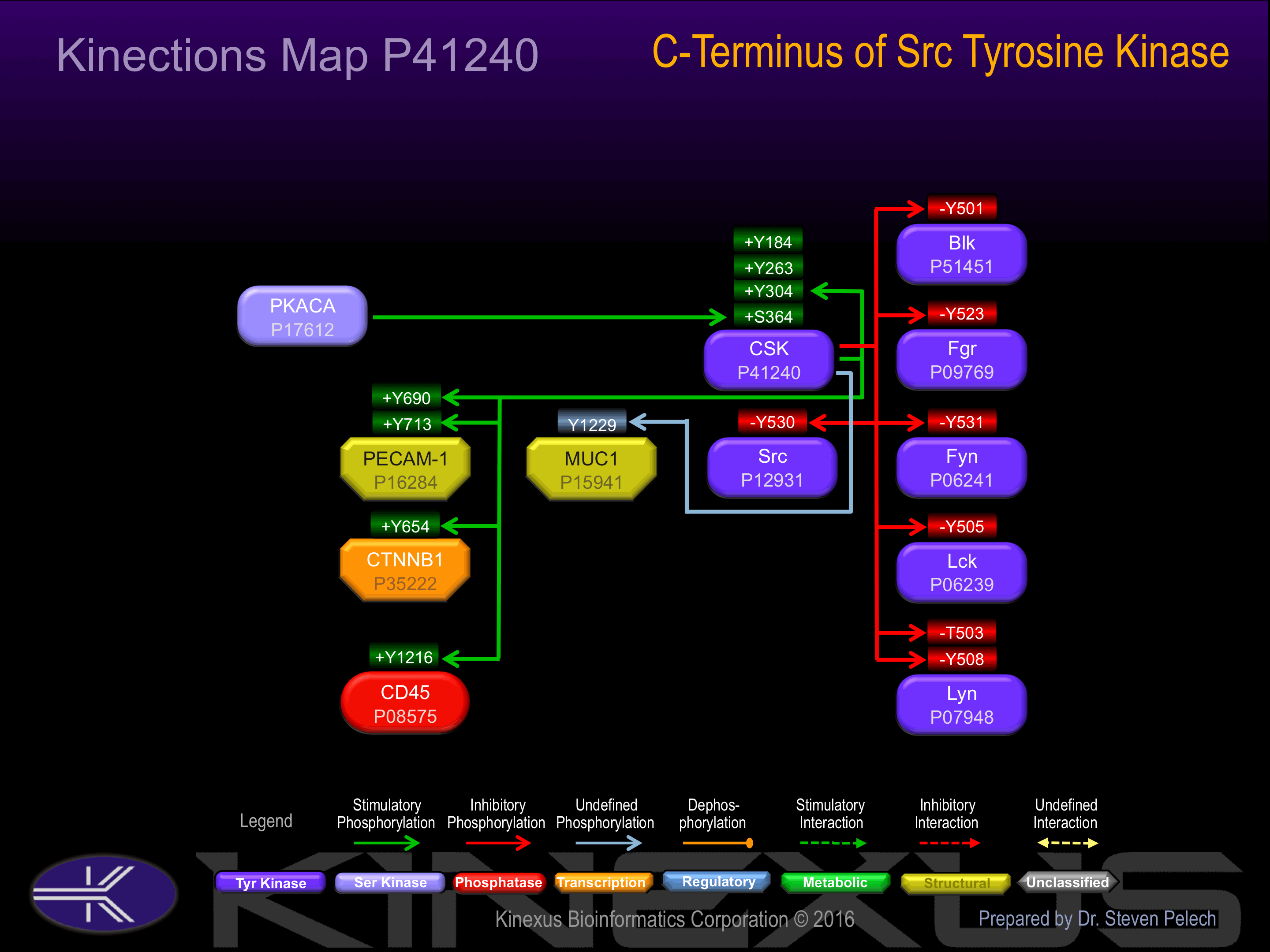
Phosphorylation of Ser-364 increases phosphotransferase activity via SH3 domain of Csk. Phosphorylation of Tyr-184 and possibly Tyr-304 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Blk | P51451 | Y501 | YTATERQYELQP___ | - |
| CD45 | P08575 | Y1216 | MVSTFEQYQFLYDVI | + |
| CSK | P41240 | Y304 | DVCEAMEYLEGNNFV | + |
| CTNNB1 | P35222 | Y654 | RNEGVATYAAAVLFR | + |
| Fgr | P09769 | Y523 | FTSAEPQYQPGDQT_ | - |
| Fyn | P06241 | Y531 | FTATEPQYQPGENL_ | - |
| Lck | P06239 | Y505 | FTATEGQYQPQP___ | - |
| Lyn | P07948 | T503 | LDDFYTATEGQYQQQ | - |
| Lyn | P07948 | Y508 | YTATEGQYQQQP___ | - |
| MUC1 | P15941 | Y1229 | SSTDRSPYEKVSAGN | ? |
| PECAM-1 | P16284 | Y690 | PLNSDVQYTEVQVSS | + |
| PECAM-1 | P16284 | Y713 | KKDTETVYSEVRKAV | + |
| Src | P12931 | Y530 | FTSTEPQYQPGENL_ | - |
| Yes | P07947 | Y537 | FTATEPQYQPGENL_ | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
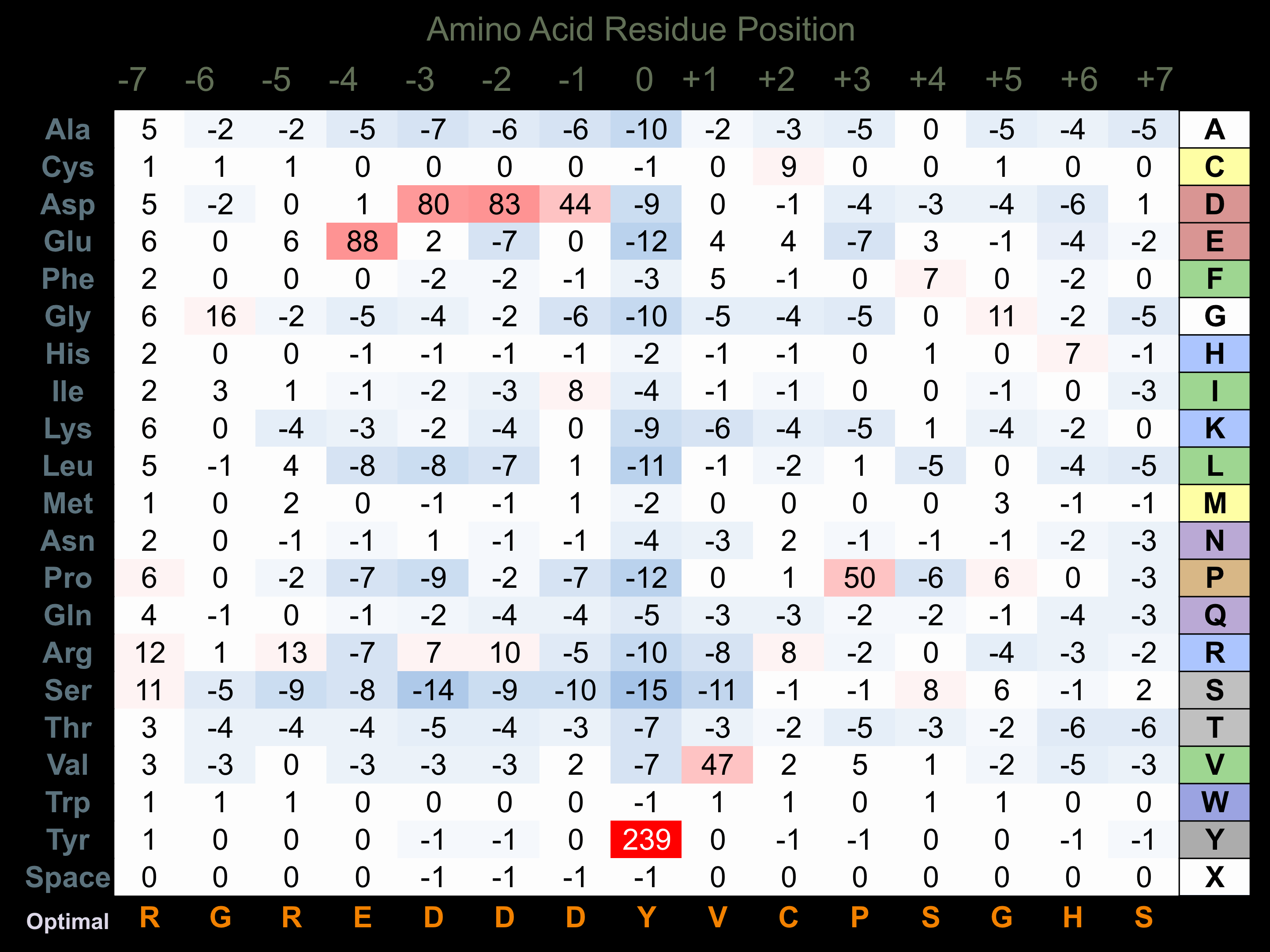
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Autoimmune disorders
Specific Diseases (Non-cancerous):
Systemic lupus erythematosus (SLE)
Comments:
CSK appears to be a tumour suppressor protein (TSP), although it seems to undergo a typical rate bystander mutations in human cancers. The active form of the protein kinase might normally acts to inhibit tumour cell proliferation through phosphorylation of the C-terminal inhibitory tyrosine of Src-related protein kinases, which represses their activity.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -76, p<(0.0003); Brain oligodendrogliomas (%CFC= -75, p<0.0002); Classical Hodgkin lymphomas (%CFC= +58, p<0.016); Large B-cell lymphomas (%CFC= +358, p<0.0002); Ovary adenocarcinomas (%CFC= +177, p<0.0006); Pituitary adenomas (ACTH-secreting) (%CFC= -60); Skin fibrosarcomas (%CFC= +82); Skin melanomas - malignant (%CFC= +234, p<0.0001); and Skin squamous cell carcinomas (%CFC= +52, p<0.093). The COSMIC website notes an up-regulated expression score for CSK in diverse human cancers of 347, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 8 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A Y184F mutation can lead to inhibited phosphorylation of CSK. It phosphotransferase activity can be decreased by two-thirds with a Y304F mutation. PRKACA induced phosphorylation of CSK can be inhibited with a CSK mutation S364A.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24434 diverse cancer specimens. This rate is only -7 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.44 % in 555 stomach cancers tested; 0.34 % in 1052 large intestine cancers tested; 0.3 % in 805 skin cancers tested; 0.14 % in 1594 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: Q26* (4).
Comments:
Only 2 deletions, no insertions or complex mutations are noted on the COSMIC website.