Nomenclature
Short Name:
DAPK3
Full Name:
Death-associated protein kinase 3
Alias:
- DAP kinase 3
- DAP-like kinase
- ZIP-kinase
- Dlk
- EC 2.7.11.1
- FLJ36473
- ZIPK
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
DAPK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
52,536
# Amino Acids:
454
# mRNA Isoforms:
2
mRNA Isoforms:
52,536 Da (454 AA; O43293); 37,048 Da (322 AA; O43293-2)
4D Structure:
Homodimer or forms heterodimers with ATF4. Both interactions require an intact leucine zipper domain and oligomerization is required for full enzymatic activity. Also binds to DAXX and PAWR, possibly in a ternary complex which plays a role in caspase activation. Interacts with AATF, CDC5L, UBE2D1, UBE2D2 AND UBE2D3
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
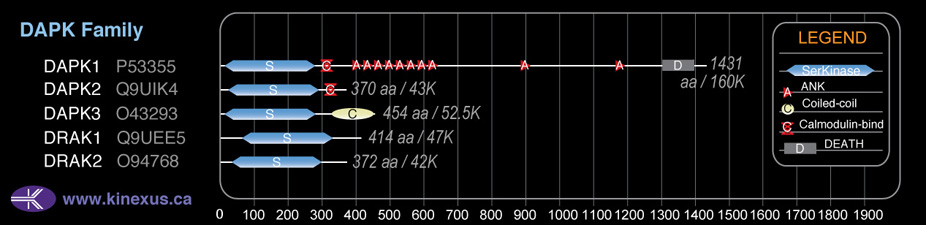
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 13 | 275 | Pkinase |
| 329 | 445 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S50, S311, S312, S407, S414.
Threonine phosphorylated:
T180-, T225+, T265+, T299+, T306, T388.
Tyrosine phosphorylated:
Y39, Y305.
Ubiquitinated:
K167, K327, K425.
Methylated:
K37.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 67
67
793
38
1126
 5
5
60
16
42
 22
22
262
9
196
 31
31
372
111
883
 56
56
660
31
599
 6
6
74
90
200
 16
16
184
41
481
 100
100
1183
37
2094
 32
32
378
17
286
 7
7
81
97
149
 7
7
87
28
61
 53
53
627
166
498
 5
5
64
31
74
 7
7
84
12
55
 13
13
156
22
172
 3
3
40
20
33
 17
17
206
120
112
 6
6
66
18
36
 6
6
66
102
69
 39
39
456
137
513
 13
13
155
20
151
 11
11
133
24
126
 23
23
271
11
220
 10
10
121
18
77
 8
8
99
20
95
 82
82
969
70
2160
 6
6
66
34
57
 10
10
122
18
87
 21
21
244
18
183
 7
7
85
42
93
 55
55
647
24
633
 52
52
621
41
856
 14
14
167
78
411
 50
50
591
78
519
 5
5
62
48
101
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 25
25
29.3
100 98.9
98.9
99.8
99 -
-
-
95 -
-
-
90 95.4
95.4
97.8
95 -
-
-
- 83.9
83.9
91
85 83.7
83.7
90.8
85 -
-
-
- 76.2
76.2
81.7
- 34.1
34.1
41
84 78.2
78.2
89.2
79.5 80.4
80.4
91
82 -
-
-
- 22.6
22.6
35.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 25.8
25.8
45.5
40 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PAWR - Q96IZ0 |
| 2 | AATF - Q9NY61 |
| 3 | ATF4 - P18848 |
| 4 | MET - P08581 |
| 5 | PRKCZ - Q05513 |
| 6 | GRB14 - Q14449 |
| 7 | DAXX - Q9UER7 |
| 8 | GRB2 - P62993 |
Regulation
Activation:
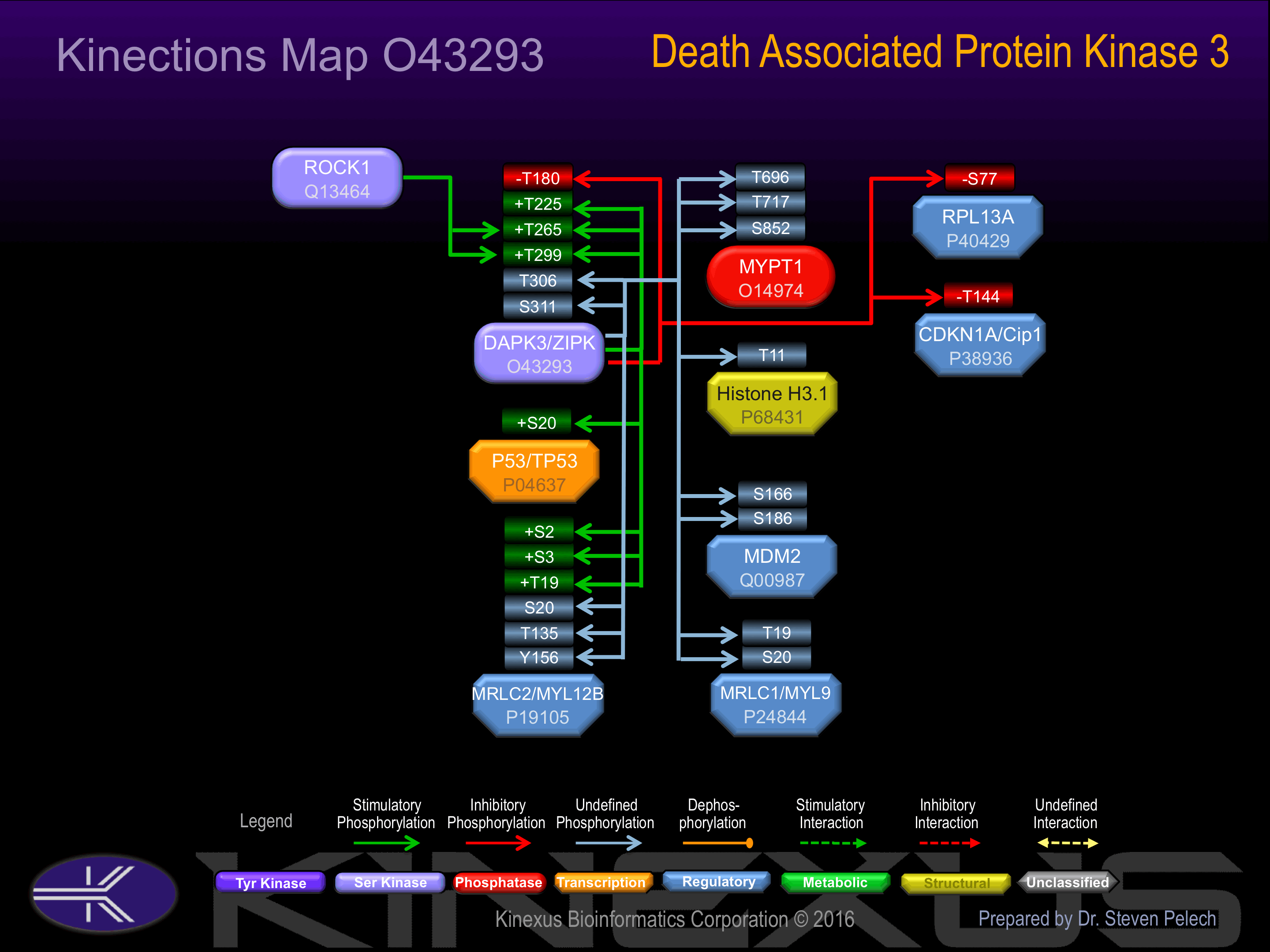
Activated by phosphorylation at Thr-180, Thr-225, and Thr-265.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Cip1 (p21, CDKN1A) | P38936 | T144 | QGRKRRQTSMTDFYH | - |
| DAPK3 | O43293 | S311 | EYTIKSHSSLPPNNS | |
| DAPK3 | O43293 | T180 | EFKNIFGTPEFVAPE | - |
| DAPK3 | O43293 | T225 | LGETKQETLTNISAV | + |
| DAPK3 | O43293 | T265 | KDPKRRMTIAQSLEH | + |
| DAPK3 | O43293 | T299 | PERRRLKTTRLKEYT | + |
| DAPK3 | O43293 | T306 | TTRLKEYTIKSHSSL | |
| H3.1 | P68431 | T11 | KQTARKSTGGKAPRK | |
| MDM2 | Q00987 | S166 | SSRRRAISETEENSD | |
| MDM2 | Q00987 | S186 | RQRKRHKSDSISLSF | |
| MRLC1 (MYL9) | P24844 | S20 | KRPQRATSNVFAMFD | |
| MRLC1 (MYL9) | P24844 | T19 | KKRPQRATSNVFAMF | |
| MRLC2 (MYL12B) | P19105 | S2 | _______SSKRTKTK | + |
| MRLC2 (MYL12B) | P19105 | S20 | KRPQRATSNVFAMFD | |
| MRLC2 (MYL12B) | P19105 | S3 | _____MSSKRTKTKT | + |
| MRLC2 (MYL12B) | P19105 | T135 | TTMGDRFTDEEVDEL | |
| MRLC2 (MYL12B) | P19105 | T19 | KKRPQRATSNVFAMF | + |
| MRLC2 (MYL12B) | P19105 | Y156 | DKKGNFNYIEFTRIL | |
| MYPT1 | O14974 | S852 | RPREKRRSTGVSFWT | |
| MYPT1 | O14974 | T696 | ARQSRRSTQGVTLTD | |
| MYPT1 | O14974 | T717 | TIGRSRSTRTREQEN | |
| p53 (TP53) | P04637 | S20 | PLSQETFSDLWKLLP | + |
| RPL13A | P40429 | S77 | PYHFRAPSRIFWRTV | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
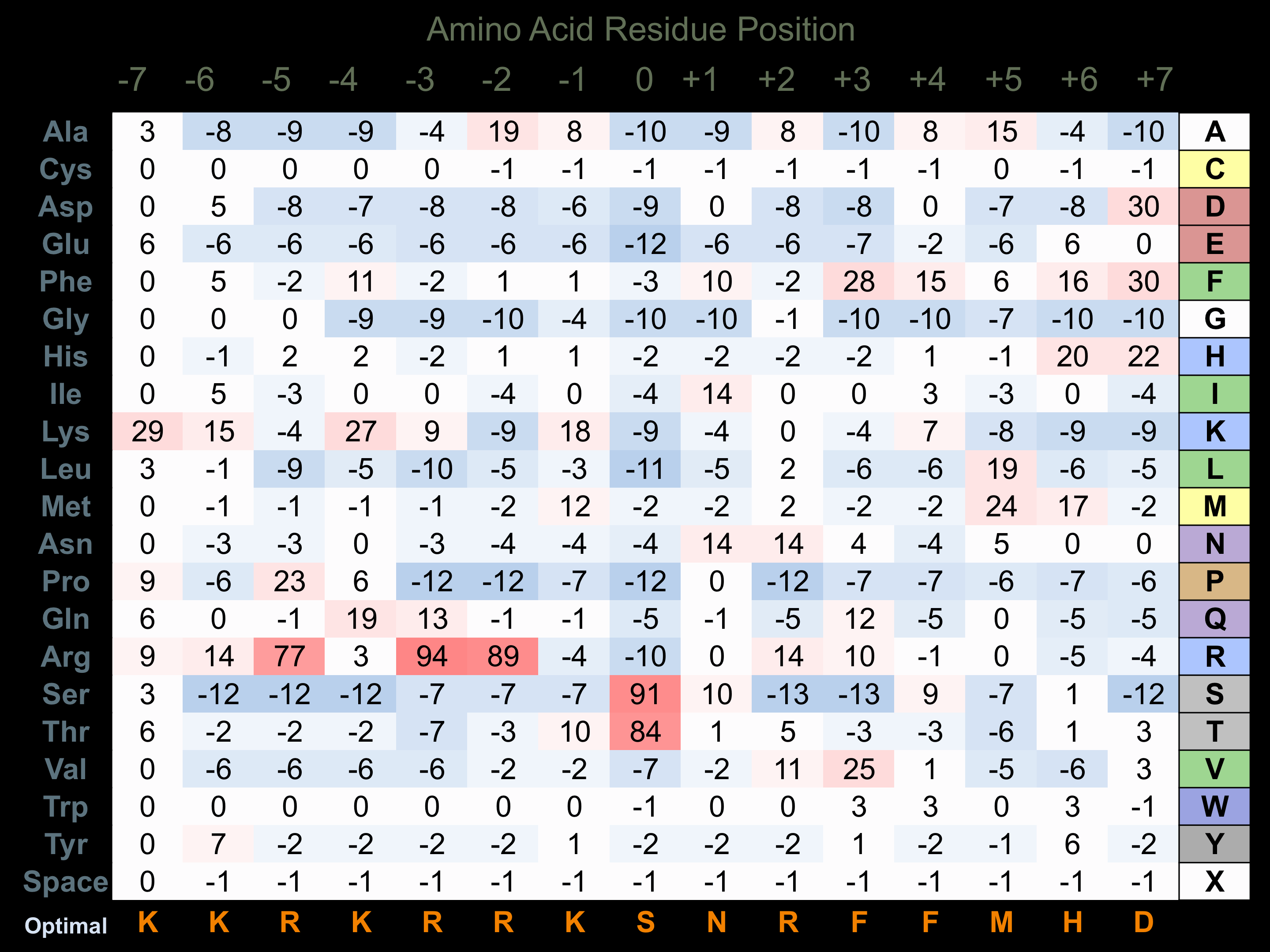
Matrix Type:
Experimentally derived from alignment of 26 known protein substrate phosphosites and 7 peptides phosphorylated by recombinant DAPK3 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, skin disorders
Specific Diseases (Non-cancerous):
Recessive dystrophic epidermolysis bullosa
Comments:
Recessive Dystrophic Epidermolysis Bullosa is an inherited disorder affecting the skin, bone, and bone marrow.
Specific Cancer Types:
Non-small cell lung cancer (NSCLC)
Comments:
DAPK3 may be a tumour suppressor protein (TSP). T112M, D161N, and P216S mutations in DAPK3 result in fully or mostly inhibited phosphotransferase activity and appear to induce cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -95, p<0.0001); and Skin melanomas - malignant (%CFC= +136, p<0.0001). The COSMIC website notes an up-regulated expression score for DAPK3 in diverse human cancers of 391, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 53 for this protein kinase in human cancers was 0.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. Examples of mutations resulting in loss of kinase phosphotransferase activity are D161A, and T225A. Loss of kinase activity will only occur at low ATP concentrations with the K42A mutation. Catalytic inhibition can occur with the mutations T265A and T299A that inhibit phosphorylation by ROCK1.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24842 diverse cancer specimens. This rate is only 18 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.49 % in 1361 large intestine cancers tested; 0.4 % in 273 cervix cancers tested; 0.33 % in 864 skin cancers tested; 0.26 % in 589 stomach cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,126 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.