Nomenclature
Short Name:
EGFR
Full Name:
Epidermal growth factor receptor
Alias:
- EC 2.7.10.1
- EGFR
- HER1
- Receptor tyrosine-protein kinase ErbB-1
- V-erb-b oncogene homologue
- Epidermal growth factor receptor
- ErbB-1
- ErbB
- mENA
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
EGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
134,277
# Amino Acids:
1210
# mRNA Isoforms:
4
mRNA Isoforms:
134,277 Da (1210 AA; P00533); 77,312 Da (705 AA; P00533-3); 69,228 Da (628 AA; P00533-4); 44,664 Da (405 AA; P00533-2)
4D Structure:
Binds RIPK1. Interacts (via autophosphorylated C-terminal tail) with CBL. Part of a complex with ERBB2 and either PIK3C2A or PIK3C2B. Interacts (autophosphorylated form) with PIK3C2B; the interaction may be indirect. Interacts with PELP1. Binds MUC1. Interacts with AP2M1. Interacts with GAB2.
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
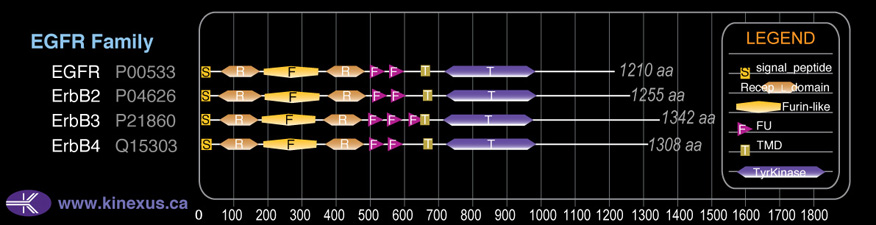
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 24 | signal_peptide |
| 57 | 168 | Recep_L_domain |
| 177 | 338 | Furin-like |
| 361 | 481 | Recep_L_domain |
| 496 | 547 | FU |
| 552 | 601 | FU |
| 646 | 668 | TMD |
| 712 | 968 | TyrKc |
| 712 | 970 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K133, K284, K346, K1179, K1182, K1188.
Methylated:
R1199 (Omega-N-methylarginine).
N-GlcNAcylated:
N56, N73, N128, N175, N196, N352, N361, N413, N444, N528, N568, N603, N623.
Other:
Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in ubiquitin) cross-linked at K716, K737, K754, K867, K929, K970. .
Serine phosphorylated:
S77, S151, S229, S511, S645, S695-, S720, S752, S768, S991-, S995, S1025, S1026-, S1030, S1036, S1037, S1039, S1042, S1045, S1057, S1064, S1070-, S1071-, S1081, S1096, S1104, S1120, S1130, S1153, S1162, S1166, S1190-.
Threonine phosphorylated:
T648, T678-, T693-, T725-, T940, T993, T1032, T1041, T1046, T1074, T1078, T1085, T1131, T1141, T1145, T1150, T1191.
Tyrosine phosphorylated:
Y112, Y113, Y117, Y270, Y585, Y727, Y764, Y801, Y827, Y869+, Y891, Y944, Y978, Y998-, Y1016+, Y1069+, Y1092+, Y1110+, Y1125, Y1138, Y1172+, Y1197+.
Ubiquitinated:
K708, K713, K714, K716, K728, K737, K739, K754, K757, K823, K846, K860, K867, K875, K913, K929, K960, K970, K1061.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 51
51
731
108
1069
 8
8
118
39
176
 17
17
248
33
255
 30
30
428
308
1029
 43
43
619
79
504
 11
11
150
246
336
 11
11
158
103
341
 100
100
1424
108
3027
 27
27
387
45
323
 6
6
80
200
98
 10
10
147
82
173
 57
57
807
449
690
 4
4
59
99
109
 5
5
68
27
61
 7
7
100
63
122
 4
4
55
52
77
 15
15
208
336
1084
 10
10
148
52
147
 6
6
83
256
89
 44
44
622
367
717
 6
6
88
58
91
 6
6
79
69
115
 9
9
126
47
122
 6
6
90
52
158
 6
6
89
58
102
 53
53
756
192
1041
 8
8
114
102
154
 12
12
176
52
157
 11
11
150
52
180
 14
14
193
84
217
 23
23
321
84
282
 47
47
671
106
850
 10
10
144
190
392
 56
56
801
213
738
 9
9
122
140
133
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.3
99.3
99.7
100 90.3
90.3
91.5
98 -
-
-
88 -
-
-
88 69.5
69.5
72.7
92 -
-
-
- 90.5
90.5
95
91 49.5
49.5
64.7
91 -
-
-
- 82.2
82.2
88.4
- 42.5
42.5
48.3
78.5 21.9
21.9
37.4
68 62.5
62.5
74
65 -
-
-
- 35.2
35.2
51.1
- -
-
-
- 27.7
27.7
46.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | EGF - P01133 |
| 2 | GRB2 - P62993 |
| 3 | SHC1 - P29353 |
| 4 | CBL - P22681 |
| 5 | PTPN1 - P18031 |
| 6 | PLCG1 - P19174 |
| 7 | SRC - P12931 |
| 8 | NCK1 - P16333 |
| 9 | TGFA - P01135 |
| 10 | ERBB2 - P04626 |
| 11 | IGHG1 - P01857 |
| 12 | NCK2 - O43639 |
| 13 | STAT3 - P40763 |
| 14 | ERRFI1 - Q9UJM3 |
| 15 | PTPN11 - Q06124 |
Regulation
Activation:
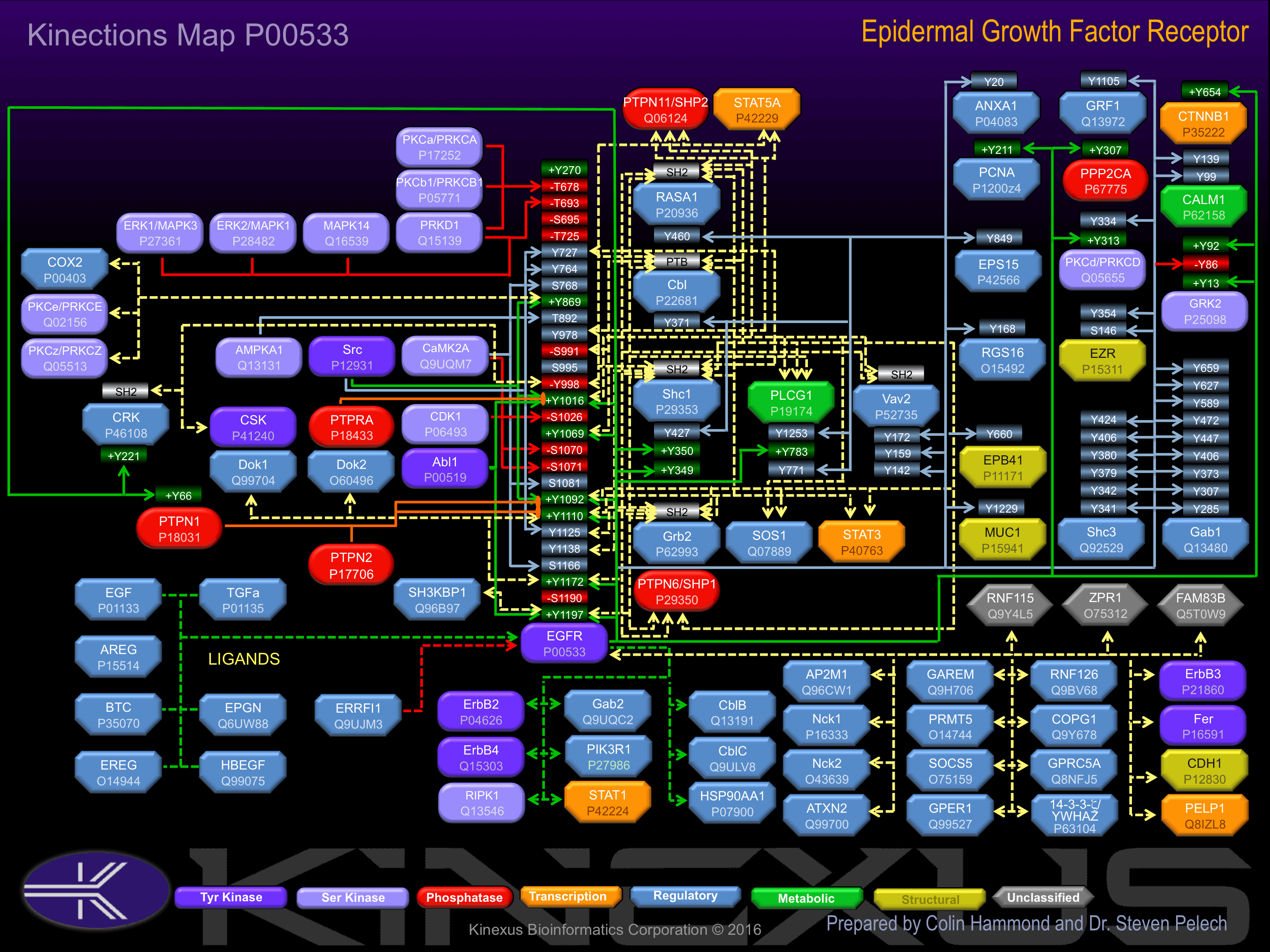
Activated by binding epidermal growth factor (EGF) and transforming growth factor-alpha (TGFa), which induces dimerization and autophosphorylation. Phosphorylation of Tyr-270 induces interaction with EGFR. Phosphorylation of Tyr-869 increases phosphotransferase activity and induces interaction with COX2. Phosphorylation of Tyr-1016 increases phosphotransferase activity and induces interaction with GrbB2, PLCg1, PTPN11 (SHP2), RasGAP, and Vav2. Phosphorylation of Tyr-1069 increases phosphotransferase activity and induces interaction with Cbl. Phosphorylation of TYr-1092 induces interaction with PLCg1, PTPN6 (SHP1) and Ras-GAP. Phosphorylation of Tyr-1110 induces interaction with Ras-GAP. Phosphorylation of Tyr-1172 increases phosphotransferase activity and induces interaction with Dok1, EGFR, RasGAP, PTPN11 (SHP2), and Vav2. Phosphorylation of Tyr-1197 increases phosphotransferase activity and induces interaction with Cbl, EGR, PLCg1, RasGAP, SH3KBP1, Shc1, and PTPN6 (SHP1).
Inhibition:
Inhibited by phosphorylation at Thr-678, Ser-695 and Ser-1026. Phosphorylation of Thr-693, Ser-695, Ser-1070, Ser-1071, Ser-1190 contributes to receptor internalization.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCb | P05771 | T678 | RHIVRKRTLRRLLQE | - |
| PKCa | P17252 | T678 | RHIVRKRTLRRLLQE | - |
| PKD1 | Q15139 | T678 | RHIVRKRTLRRLLQE | - |
| ERK2 | P28482 | T693 | RELVEPLTPSGEAPN | - |
| ERK1 | P27361 | T693 | RELVEPLTPSGEAPN | - |
| p38a | Q16539 | T693 | RELVEPLTPSGEAPN | - |
| PKD1 | Q15139 | T693 | RELVEPLTPSGEAPN | - |
| SRC | P12931 | Y727 | SGAFGTVYKGLWIPE | |
| SRC | P12931 | Y764 | KEILDEAYVMASVDN | |
| CaMK2a | Q9UQM7 | S768 | DEAYVMASVDNPHVC | |
| SRC | P12931 | Y869 | LGAEEKEYHAEGGKV | + |
| EGFR | P00533 | Y1016 | DVVDADEYLIPQQGF | + |
| SRC | P12931 | Y1016 | DVVDADEYLIPQQGF | + |
| ABL | P00519 | Y1016 | DVVDADEYLIPQQGF | + |
| CDK1 | P06493 | S1026 | PQQGFFSSPSTSRTP | - |
| EGFR | P00533 | Y1069 | EDSFLQRYSSDPTGA | + |
| CaMK2a | Q9UQM7 | S1070 | DSFLQRYSSDPTGAL | - |
| CaMK2a | Q9UQM7 | S1071 | SFLQRYSSDPTGALT | - |
| CaMK2a | Q9UQM7 | S1081 | TGALTEDSIDDTFLP | |
| EGFR | P00533 | Y1092 | TFLPVPEYINQSVPK | + |
| SRC | P12931 | Y1092 | TFLPVPEYINQSVPK | + |
| EGFR | P00533 | Y1110 | GSVQNPVYHNQPLNP | + |
| SRC | P12931 | Y1125 | APSRDPHYQDPHSTA | |
| CaMK2a | Q9UQM7 | S1166 | QKGSHQISLDNPDYQ | |
| EGFR | P00533 | Y1172 | ISLDNPDYQQDFFPK | + |
| EGFR | P00533 | Y1197 | STAENAEYLRVAPQS | + |
| ABL | P00519 | Y1197 | STAENAEYLRVAPQS | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ANXA1 | P04083 | Y20 | IENEEQEYVQTVKSS | |
| BARK1 (GRK2, ADRBK1) | P25098 | Y13 | AVLADVSYLMAMEKS | + |
| BARK1 (GRK2, ADRBK1) | P25098 | Y86 | ARPLVEFYEEIKKYE | - |
| BARK1 (GRK2, ADRBK1) | P25098 | Y92 | FYEEIKKYEKLETEE | + |
| Calmodulin | P62158 | Y139 | DGDGQVNYEEFVQMM | |
| Calmodulin | P62158 | Y99 | FDKDGNGYISAAELR | |
| Cbl | P22681 | Y371 | TQEQYELYCEMGSTF | |
| CCDC50 iso2 | Q8IVM0-2 | Y217 | MAEEKKAYKKAKERE | |
| CCDC50 iso2 | Q8IVM0-2 | Y279 | TDGEDADYTHFTNQQ | |
| CCDC50 iso2 | Q8IVM0-2 | Y304 | SSHKGFHYKH_____ | |
| Crk | P46108 | Y221 | GGPEPGPYAQPSVNT | + |
| CTNNB1 | P35222 | Y654 | RNEGVATYAAAVLFR | + |
| EGFR | P00533 | Y1016 | DVVDADEYLIPQQGF | + |
| EGFR | P00533 | Y1069 | EDSFLQRYSSDPTGA | + |
| EGFR | P00533 | Y1092 | TFLPVPEYINQSVPK | + |
| EGFR | P00533 | Y1110 | GSVQNPVYHNQPLNP | + |
| EGFR | P00533 | Y1172 | ISLDNPDYQQDFFPK | + |
| EGFR | P00533 | Y1197 | STAENAEYLRVAPQS | + |
| EPB41 | P11171 | Y660 | RLDGENIYIRHSNLM | |
| Eps15 | P42566 | Y849 | NFANFSAYPSEEDMI | |
| Ezrin | P15311 | Y146 | KEVHKSGYLSSERLI | |
| Ezrin | P15311 | Y354 | LMLRLQDYEEKTKKA | |
| GAB1 | Q13480 | Y285 | TEADGELYVFNTPSG | |
| GAB1 | Q13480 | Y307 | MRHVSISYDIPPTPG | |
| GAB1 | Q13480 | Y373 | ASDTDSSYCIPTAGM | |
| GAB1 | Q13480 | Y406 | DASSQDCYDIPRAFP | |
| GAB1 | Q13480 | Y447 | SEELDENYVPMNPNS | |
| GAB1 | Q13480 | Y472 | EPIQEANYVPMTPGT | |
| GAB1 | Q13480 | Y589 | SHDSEENYVPMNPNL | |
| GAB1 | Q13480 | Y627 | KGDKQVEYLDLDLDS | |
| GAB1 | Q13480 | Y659 | VADERVDYVVVDQQK | |
| GRF1 | Q9NRY4 | Y1105 | RNEEENIYSVPHDST | |
| MUC1 | P15941 | Y1229 | SSTDRSPYEKVSAGN | ? |
| PCNA | P12004 | Y211 | QLTFALRYLNFFTKA | + |
| PKCd (PRKCD) | Q05655 | Y313 | SSEPVGIYQGFEKKT | + |
| PKCd (PRKCD) | Q05655 | Y334 | MQDNSGTYGKIWEGS | ? |
| PKIA | P61925 | Y7 | TDVETTYADFIASG_ | |
| PLCG1 | P19174 | Y1253 | EGSFESRYQQPFEDF | |
| PLCG1 | P19174 | Y771 | IGTAEPDYGALYEGR | |
| PLCG1 | P19174 | Y783 | EGRNPGFYVEANPMP | + |
| PLD2 | O14939 | T11 | TPESLFPTGDELDSS | |
| PPP2CA | P67775 | Y307 | VTRRTPDYFL_____ | + |
| PTP1B | P18031 | Y66 | LHQEDNDYINASLIK | + |
| RasGAP | P20936 | Y460 | TVDGKEIYNTIRRKT | |
| RGS16 | O15492 | Y168 | TLMEKDSYPRFLKSP | |
| Shc1 | P29353 | Y349 | EEPPDHQYYNDFPGK | + |
| Shc1 | P29353 | Y350 | EPPDHQYYNDFPGKE | + |
| Shc1 | P29353 | Y427 | ELFDDPSYVNVQNLD | ? |
| Shc3 | Q92529 | Y341 | GDGSDHPYYNSIPSK | ? |
| Shc3 | Q92529 | Y342 | DGSDHPYYNSIPSKM | ? |
| Shc3 | Q92529 | Y379 | FAGKEQTYYQGRHLG | |
| Shc3 | Q92529 | Y380 | AGKEQTYYQGRHLGD | |
| Shc3 | P29353 | Y406 | RQGSSDIYSTPEGKL | |
| Shc3 | P29353 | Y424 | PTGEAPTYVNTQQIP | |
| VAV2 | P52735 | Y142 | TENDDDVYRSLEELA | |
| VAV2 | P52735 | Y159 | HDLGEDIYDCVPCED | |
| VAV2 | P52735 | Y172 | EDGGDDIYEDIIKVE |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
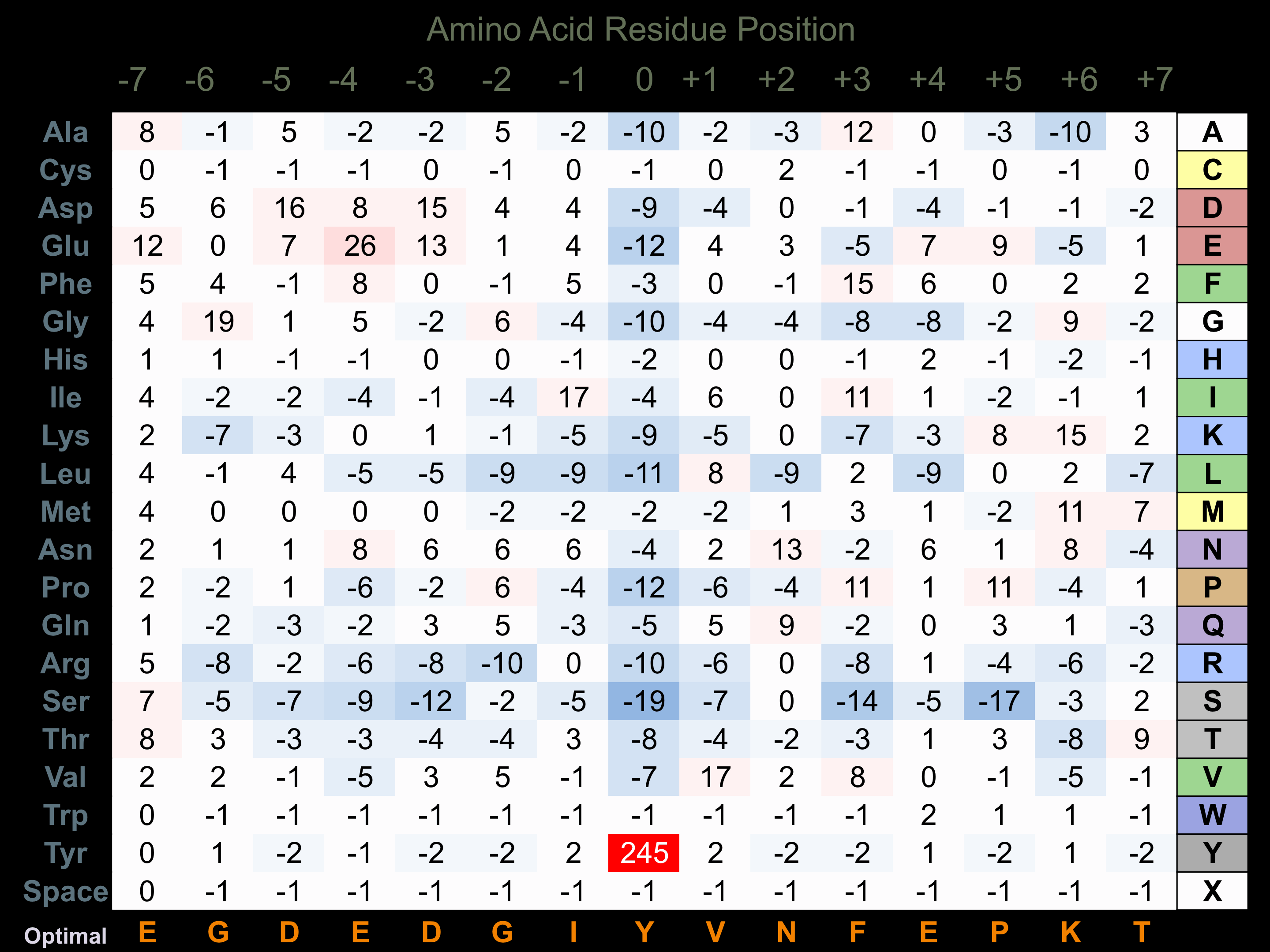
Matrix Type:
Experimentally derived from alignment of 78 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, immune, skin, heart, gastrointestinal, fetal, and neuronal disorders
Specific Diseases (Non-cancerous):
Paronychia; Folliculitis; Exanthem (RASH); Left ventricular outflow tract obstruction; Colonic disease; Placenta accreta; Troyer syndrome; Polycystic kidney disease, Type 1
Comments:
Paronychia is related to lung cancer and is characterized by bacterial or fungal infections in the nails. Folliculitis can affect bone, bone marrow, and skin. Folliculitis can affect one or more follicles with an inflammatory response on anywhere but the feet or hands. Exanthem is irritated or swollen skin arising from contact with an irritant. Left Ventricular Outflow tract Obstruction is a hypertensive heart disease that can also affect liver and lung tissues. Colonic Disease can include ulcerative colitis (colon ulcers), rectum diverticulitis (inflammation leading to irritable bowel syndrome), and colorectal cancer colonic polyps. Placenta Accreta arises from the placenta attaching itself to the myometrium rather than endometrium. Placenta Accreta can lead to sepsis. Troyer Syndrome is a rare neuronal disease that induces muscle stiffness, leading to paralysis of the lower body. Polycystic Kidney Disease, Type 1 (PKD1) is related to the cystic kidney disorder.
Specific Cancer Types:
Lung cancer (LC), EGFR-related; Lung cancer (LC); Breast cancer; Pancreatic cancer; Glioblastoma multiforme; Adenocarcinomas; Astrocytomas; Giant cell glioblastomas (GBM); Prostate cancer; Non-small cell lung carcinomas (NSCLC); Oligodendroglioma; leukemias; Brain cancer; Paronychia; Colon cancer; Renal cell carcinomas (RCC); Adenosquamous carcinomas; Pulmonary blastomas; Malignant peritoneal mesotheliomas; Blastomas; Adenocarcinomas in Situ; Lip cancer (lower); Sinonasal undifferentiated carcinomas (SNUC); Cervical adenosquamous carcinomas; Ring chromosome 7 (R7); Transitional cell carcinomas (TCC); Primary peritoneal carcinomas (PPC); Sweat gland carcinomas; Placental site trophoblastic tumours (PSST); Pleomorphic xanthoastrocytomas (PXA); Thymic epithelial neoplasms (TEN); Lymphoepithelioma-like carcinomas (LELC); Mammary Paget's disease; Hidradenomas; Colonic disease; Pancreatic endocrine carcinomas; Hidradenocarcinomas; Intraocular melanomas; Nonpapillary renal cell carcinomas; Microglandular adenosis is a breast cancer; Congenital pulmonary airway Malformation (CCAM); Basaloid squamous cell carcinomas; Gemistocytic astrocytomas; Superior vena cava syndrome (SVCS); Papillary glioneuronal tumours (PGNT); Brain stem gliomas; Ethmoid sinus adenocarcinomas; Lung cancer (LC) susceptibility; Orbital lymphangiomas; Spinal chordomas; Esophageal basaloid squamous cell carcinomas; Brain ependymomas; Gliosarcomas; Lung adenocarcinomas
Comments:
EGFR is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. EGFR mutations have been linked to Glioblastoma Multiforme, which is related to astrocytoma and anaplastic astrocytoma, with regions of anaplastic cells surrounding necrotizing tissue. Glioblastoma Multiforme affects brain, endothelial, and bone tissues. Astrocytoma is the development of cancer arising from a glial cell, typically in the cerebrum, but often in the spinal cord as well. Oligodendroglioma can affect brain, spinal cord, and endothelial tissues and is a cancer arising from oligodendrocytes. Paronychia is related to lung cancer and is characterized by bacterial or fungal infections in the nails. Adenosquamous Carcinoma is a cancer that can arise from squamous cells and gland-like cells. Pulmonary Blastoma can arise either from lung tissue or from the pleura tissue which lines the chest cavity, covering the lungs. Malignant Peritoneal Mesothelioma is rare, and is characterized by edema, or collection of watery fluids in the lower limbs. Malignant Peritoneal Mesothelioma can affect the lung, colon, and lymph nodes as well. Sinonasal Undifferentiated Carcinoma is a rare cancer that develops in the sinus from rapid expansion of epithelial tissue. SNUC can lead to blocked breathing. Ring Chromosome 7 (R7) is a rare disease, and involves ERK signalling and PI-3K cascade signalling pathways in its development of disease. R7 can affect T cells, heart, and skin. Transitional Cell Carcinoma (TCC) is a cancer of the urinary system, affecting the kidney, urinary bladder, prostate, and ovary. Primary Peritoneal Carcinoma is a cancer of the peritoneal cavity and can affect the ovary, endothelial, and breast tissues. Sweat Gland Carcinoma is a tumour arising from sweat gland cells and can affect breast, skin, and lymph node tissues. Placental Site Trophoblastic tumour can induce spontaneous abortions (miscarriages), and is a gestational trophoblastic disease. Pleomorphic Xanthoastrocytoma is a rare brain cancer that primarily affects children and teenagers. Thymic Epithelial Neoplasm (TEN) is a cancer related to breast cancer and thymic epithelial tumours. TEN is a rare disease and can affect thymus, lung, and endothelial tissues. Lymphoepithelioma-Like Carcinoma is a tumour arising from the endothelium. Mammary Paget's Disease is a rare cancer of the nipple. Hidradenoma are benign tumours of sweat glands that can also affect skin, breast, and salivary gland tissues. Colonic Disease can include ulcerative colitis (colon ulcers), rectum diverticulitis (inflammation leading to irritable bowel syndrome), and colorectal cancer colonic polyps. Hidradenocarcinoma is a rare disease arising from unregulated tissue growth in sweat glands and development of a tumour. Intraocular Melanoma is a rare eye cancer arising from the pigment-producing cells (melanocytes) which reside in the uveal tract. Congenital Pulmonary Airway Malformation is related to mucoepidermoid carcinoma and adenocarcinoma and can lead to lung failure. Basaloid Squamous Cell Carcinoma is a cancer of the squamous cells (outer layer of skin). Gemistocytic Astrocytoma is a rare cancer related to astrocytoma and subependymal giant cell astrocytoma. Superior Vena Cava Syndrome (SCVS) is a rare disease obstructing blood flow into the superior vena cava (artery supplying the upper portion of the heart), and this disease is related to lung and breast cancer. Papillary Glioneuronal tumour is related to neuronitis and siderosis. Ethmoid Sinus Adenocarcinoma is related to the inverted papilloma and adenocarcinoma diseases. Orbital Lymphangioma is a rare lymphoma that can affect retinal and endothelial tissue. Brain Ependymoma is a neuronal cancer disease. Gliosarcoma is also a neuronal cancer disease, though it is rare and it arises from glial cells. lung Adenocarcinoma is a rare cancer partly characterized by its large production of mucus and which is associated with adenocarcinoma and lung cancer. Tissues affected by lung Adenocarcinoma include lung, brain, and endothelium.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -76, p<0.0004); Brain oligodendrogliomas (%CFC= -96, p<0.0001); Breast epithelial carcinomas (%CFC= -56, p<0.011); Clear cell renal cell carcinomas (cRCC) (%CFC= +291, p<0.012); Head and neck squamous cell carcinomas (HNSCC) (%CFC= +118, p<0.006); Ovary adenocarcinomas (%CFC= +367, p<0.006); Prostate cancer - primary (%CFC= +103, p<0.0001); and Skin melanomas - malignant (%CFC= -69, p<(0.0003). The COSMIC website notes an up-regulated expression score for EGFR in diverse human cancers of 681, which is 1.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 4 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. Autophosphorylation of the EGFR can be fully inhibited with a R429E mutation. Autophosphorylation of the EGFR can be reduced with the Y275A (when associated with R309A), F287A (when associated with R309A), R309S (when associated with Y275A and F287A), and V689A mutations. Phosphorylation of EGFR can be fully inhibited with any of the N700A, L704A, R705A, I706A, and K745AM mutations. Reduced phosphorylation of EGFR can occur with the L688A, E690A, L692A/P, T693D, P694A, P699A, D974A, and R977A mutations. Increased phosphorylation of EGFR can occur with a T693A mutation. EGF binding to EGFR is promoted with the D587A+H590A, or K609A mutations. Constitutive activation of EGFR leading to constitutive downstream kinase activity has been seen with the mutations of V689M, E1005R+D1006K. A 50% decrease in interaction with PIK3C2B has been noted with an EGFR mutation of Y1016F. The interaction is decreased by 65% when the Y1016F mutation is associated with Y1197F. This interaction is fully inhibited with the mutations resulting in Y1197F and Y1092F. The Y1092F mutation alone will not affect the EGFR interaction with PIK3C2B, but will fully inhibit interactions when associated with F-1197 and F-1016 mutations. The Y1197F mutation alone will not affect the EGFR interaction with PIK3C2B, but when associated with Y1016F the EGFR-PIK3C2B interaction is inhibited by 65%, and when associated with Y1016F and Y1092F the interaction is fully inhibited. Interaction between EGFR and CBLC is heavily impaired with a R1068G mutation, and fully impaired with an Y1069F mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 1.53 % in 130410 diverse cancer specimens. This rate is 20.4-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 2.36 % in 80494 lung cancers tested; 0.64 % in 142 peritoneum cancers tested; 0.52 % in 3730 central nervous system cancers tested; 0.33 % in 894 endometrium cancers tested; 0.33 % in 5136 large intestine cancers tested; 0.32 % in 1577 stomach cancers tested; 0.28 % in 1304 prostate cancers tested; 0.27 % in 2102 skin cancers tested; 0.18 % in 688 biliary tract cancers tested; 0.18 % in 3551 upper aerodigestive tract cancers tested; 0.17 % in 2160 oesophagus cancers tested; 0.17 % in 1822 ovary cancers tested; 0.13 % in 834 urinary tract cancers tested; 0.09 % in 4418 breast cancers tested; 0.09 % in 2966 haematopoietic and lymphoid cancers tested; 0.09 % in 1933 kidney cancers tested; 0.09 % in 1452 thyroid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: L858R (9181); T790M (1093); G719A (166); G719S (115).
Comments:
Over 3000 deletions mutations at E746, over 100 insertion mutations clustered between D770 and H773, and over 600 complex mutations around E746 and L747 are noted on the COSMIC website. These mutations are clustered in the first quarter of the kinase catalytic domain of the EGFR.