Nomenclature
Short Name:
PKD1
Full Name:
Protein kinase C, mu type
Alias:
- EC 2.7.11.13
- PKC-mu
- PKD
- PRKCM
- PRKD1
- Protein kinase D1; Protein kinase C, mu type; Protein kinase D;
- Kinase PKD1
- KPCD1
- NPKC-mu
- PKCM
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
PKD
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
101704
# Amino Acids:
912
# mRNA Isoforms:
1
mRNA Isoforms:
101,704 Da (912 AA; Q15139)
4D Structure:
Interacts (via N-terminus) with ADAP1/CENTA1. Interacts with Src
1D Structure:
Subfamily Alignment
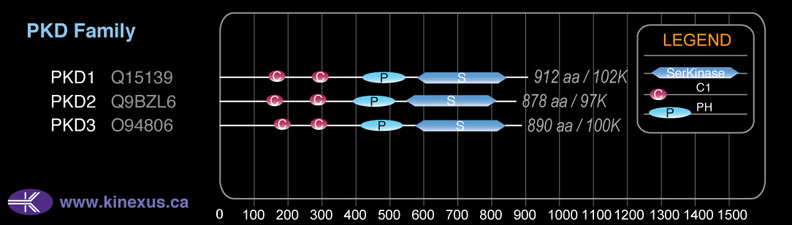
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K612.
Serine phosphorylated:
S205-, S208-, S219-, S223-, S225, S235, S237, S239, S247, S249+, S345, S379, S397-, S401, S412, S421, S448, S460, S473, S548, S549, S738+, S742+, S890, S892, S910+.
Threonine phosphorylated:
T210, T217, T224, T395+, T422, T746-.
Tyrosine phosphorylated:
Y95+, Y432, Y443, Y463+, Y502, Y749-, .
Ubiquitinated:
K292, K737.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
590
29
788
 8
8
50
15
29
 17
17
103
19
166
 32
32
187
98
327
 88
88
518
25
472
 56
56
329
82
1432
 25
25
145
31
348
 76
76
446
47
871
 54
54
320
17
259
 18
18
106
90
121
 7
7
40
37
92
 89
89
526
170
533
 10
10
58
41
177
 14
14
83
12
72
 8
8
45
15
33
 9
9
55
15
61
 15
15
88
129
140
 9
9
52
28
79
 5
5
32
98
31
 62
62
363
109
405
 18
18
106
30
94
 11
11
65
33
187
 11
11
65
29
116
 21
21
124
28
127
 10
10
61
30
149
 90
90
530
64
940
 5
5
28
44
76
 13
13
74
28
169
 17
17
98
28
114
 13
13
78
28
55
 42
42
248
24
247
 60
60
355
36
367
 34
34
202
67
415
 90
90
532
57
515
 24
24
144
35
133
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.4
99.4
99.5
99.5 98.9
98.9
99.3
99 -
-
-
97 -
-
-
99 92
92
93.9
97 -
-
-
- 92.5
92.5
95
93.5 93.1
93.1
95.4
94 -
-
-
- 81.3
81.3
86.3
- -
-
-
90 -
-
-
84 80.1
80.1
86.7
83 -
-
-
- 20.1
20.1
32
- -
-
-
- 43.7
43.7
56.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20.2
20.2
34.4
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PRKCE - Q02156 |
| 2 | HDAC5 - Q9UQL6 |
| 3 | CDH1 - P12830 |
| 4 | BTK - Q06187 |
| 5 | MT2A - P02795 |
| 6 | EGFR - P00533 |
| 7 | HDAC7 - Q8WUI4 |
| 8 | KIDINS220 - Q9ULH0 |
| 9 | IGF1R - P08069 |
| 10 | C1QBP - Q07021 |
| 11 | PPP1R14A - Q96A00 |
| 12 | YWHAZ - P63104 |
| 13 | COX19 - Q49B96 |
| 14 | ADAP1 - O75689 |
Regulation
Activation:
Activated by DAG and phorbol esters. Phorbol-ester/DAG-type domain 1 binds DAG with high affinity and appears to play the dominant role in mediating translocation to the cell membrane and trans-Golgi network. Phorbol-ester/DAG-type domain 2 binds phorbol ester with higher affinity. Phosphorylation at Tyr-95 increases phosphotransferase activity and induces interaction with PKC-delta.Phosphorylation at Ser-249 and Tyr-463 increases phosphotransferase activity. Phosphorylation at Ser-738 and Ser-742 increases phosphotransferase activity and induces interaction with ASK1, JNK1 and IKKb. Phosphorylation at Ser-910 increases phosphotransferase activity and prolongs kinase activation.
Inhibition:
Autophosphorylation of Ser-742 and phosphorylation of Ser-738 by PKC relieves auto-inhibition by the PH domain. Phosphorylation at Ser-205, Ser-208, Ser-219 and Ser-223 induces binding of 14-3-3 beta and tau to inhibit PKC-mu phosphotransferase activity. Phosphorylation at Ser-397 induces interaction with 14-3-3 beta.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| SRC | P12931 | Y95 | KFPECGFYGMYDKIL | + |
| PKD1 | Q15139 | S205 | GVRRRRLSNVSLTGV | - |
| p38d | O15264 | S205 | GVRRRRLSNVSLTGV | - |
| p38d | O15264 | S208 | RRRLSNVSLTGVSTI | - |
| p38d | O15264 | S249 | GREKRSNSQSYIGRP | + |
| PKCa | P17252 | S249 | GREKRSNSQSYIGRP | + |
| p38d | O15264 | S397 | EDANRTISPSTSNNI | - |
| p38d | O15264 | S401 | RTISPSTSNNIPLMR | |
| ABL | P00519 | Y432 | KEGWMVHYTSKDTLR | |
| SRC | P12931 | Y432 | KEGWMVHYTSKDTLR | |
| ABL | P00519 | Y463 | NDTGSRYYKEIPLSE | + |
| SRC | P12931 | Y463 | NDTGSRYYKEIPLSE | + |
| ABL | P00519 | Y502 | TTANVVYYVGENVVN | |
| SRC | P12931 | Y502 | TTANVVYYVGENVVN | |
| PKCh | P24723 | S738 | ARIIGEKSFRRSVVG | + |
| PKCd | Q05655 | S738 | ARIIGEKSFRRSVVG | + |
| PKCe | Q02156 | S738 | ARIIGEKSFRRSVVG | + |
| PKD1 | Q15139 | S738 | ARIIGEKSFRRSVVG | + |
| PKD3 | O94806 | S738 | ARIIGEKSFRRSVVG | + |
| PKCz | Q05513 | S738 | ARIIGEKSFRRSVVG | + |
| PKCa | P17252 | S738 | ARIIGEKSFRRSVVG | + |
| PKCh | P24723 | S742 | GEKSFRRSVVGTPAY | + |
| PKCd | Q05655 | S742 | GEKSFRRSVVGTPAY | + |
| PKCe | Q02156 | S742 | GEKSFRRSVVGTPAY | + |
| PKD1 | Q15139 | S742 | GEKSFRRSVVGTPAY | + |
| PKD3 | O94806 | S742 | GEKSFRRSVVGTPAY | + |
| PKCz | Q05513 | S742 | GEKSFRRSVVGTPAY | + |
| p38d | O15264 | S742 | GEKSFRRSVVGTPAY | + |
| PKCa | P17252 | S742 | GEKSFRRSVVGTPAY | + |
| PKD1 | Q15139 | S910 | KALGERVSIL_____ | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| Akt1 (PKBa) | P31749 | T308 | KDGATMKTFCGTPEY | + |
| AP-2 alpha | P05549 | S258 | GVLRRAKSKNGGRSL | |
| AP-2 alpha | P05549 | S326 | EFLNRQHSDPNEQVT | |
| Cofilin 1 | P23528 | S2 | ______ASGVAVSDG | |
| COL4A3BP | Q9Y5P4 | S132 | SSLRRHGSMVSLVSG | - |
| Cortactin | Q14247 | S298 | EKLAKHESQQDYSKG | |
| Cortactin | Q14247 | S348 | EAVTSKTSNIRANFE | |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| CREB1 | P16220 | S98 | KDLKRLFSGTQISTI | |
| CTNNB1 | P35222 | T112 | EGMQIPSTQFDAAHP | |
| CTNNB1 | P35222 | T120 | QFDAAHPTNVQRLAE | |
| HDAC5 | Q9UQL6 | S259 | FPLRKTASEPNLKVR | |
| HDAC5 | Q9UQL6 | S498 | RPLSRTQSSPLPQSP | |
| HDAC7 | Q8WUI4 | S155 | FPLRKTVSEPNLKLR | |
| HDAC7 | Q8WUI4 | S358 | WPLSRTRSEPLPPSA | |
| HDAC7 | Q8WUI4 | S486 | RPLSRAQSSPAAPAS | |
| HPK1 (MAP4K1) | Q92918 | S171 | ATLARRLSFIGTPYW | + |
| HSP27 | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| Jun (c-Jun) | P05412 | S63 | KNSDLLTSPDVGLLK | + |
| Kidins220 | Q9ULH0 | S924 | RTITRQMSFDLTKLL | |
| MARK2 | Q7KZI7 | S400 | HKVQRSVSANPKQRR | ? |
| PIK4CB | Q9UBF8 | S294 | SNLKRTASNPKVENE | + |
| PIP5K2A | P48426 | T376 | KAAHAAKTVKHGAGA | - |
| PKD1 (PRKCM) | Q15139 | S205 | GVRRRRLSNVSLTGV | - |
| PKD1 (PRKCM) | Q15139 | S738 | ARIIGEKSFRRSVVG | + |
| PKD1 (PRKCM) | Q15139 | S742 | GEKSFRRSVVGTPAY | + |
| PKD1 (PRKCM) | Q15139 | S910 | KALGERVSIL_____ | + |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PTRH2 | Q9Y3E5 | S5 | ___MPSKSLVMEYLA | |
| PTRH2 | Q9Y3E5 | S87 | GKVAAQCSHAAVSAY | |
| Rin1 | Q13671 | S351 | RPLLRSMSAAFCSLL | - |
| SSH1 | Q8WYL5 | S978 | SPLKRSHSLAKLGSL | - |
| TLR5 | O60602 | S805 | YQLMKHQSIRGFVQK | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| VR1 | Q8NER1 | S117 | LRLYDRRSIFEAVAQ | |
| EGFR | P00533 | T678 | RHIVRKRTLRRLLQE | - |
| EGFR | P00533 | T693 | RELVEPLTPSGEAPN | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
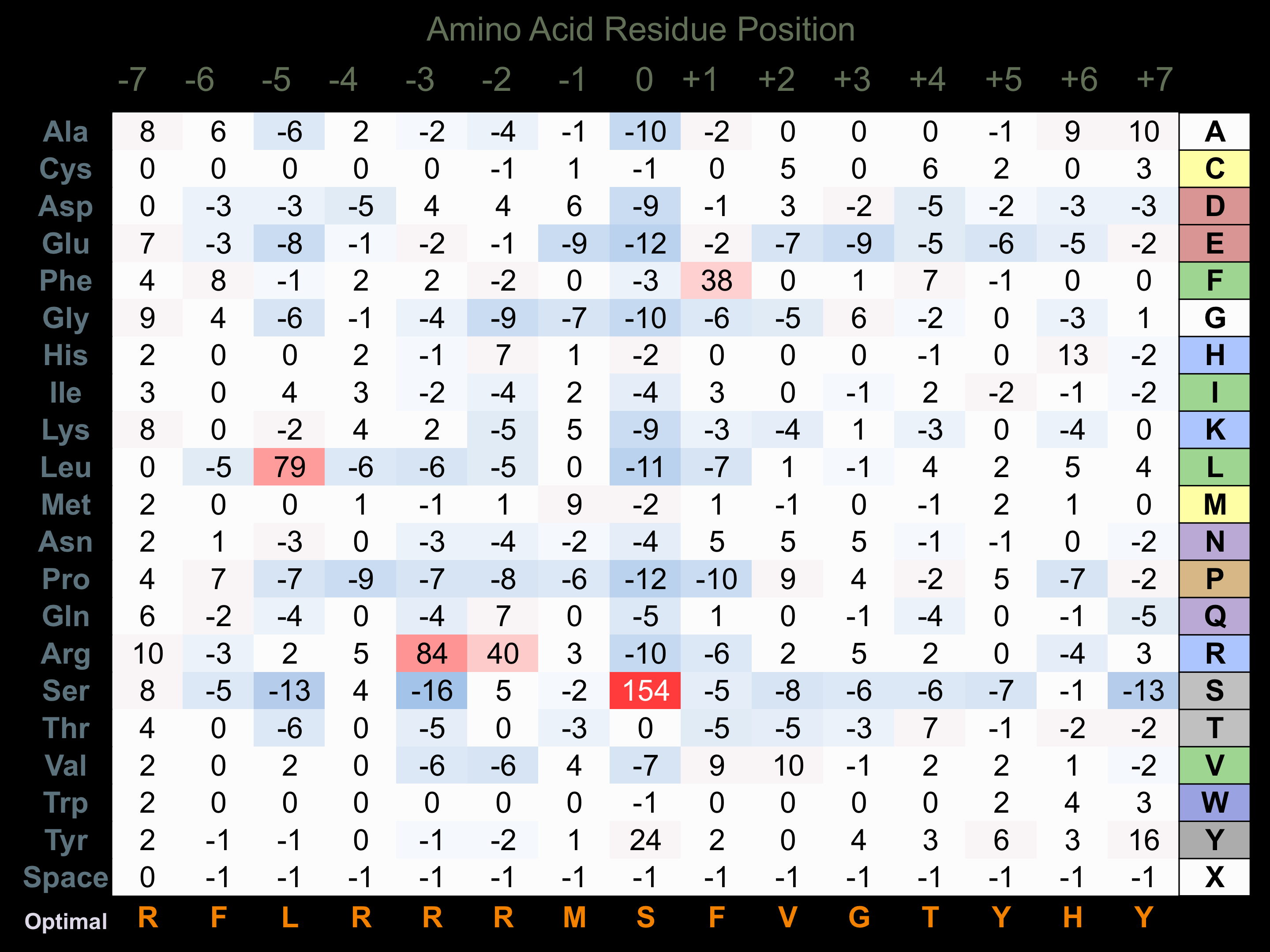
Matrix Type:
Experimentally derived from alignment of 68 known protein substrate phosphosites and 25 peptides phosphorylated by recombinant PKD1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Gastric cancer (%CFC= -53, p<0.056); Ovary adenocarcinomas (%CFC= -50, p<0.047); and Prostate cancer (%CFC= -45, p<0.098). The COSMIC website notes an up-regulated expression score for PKD1 in diverse human cancers of 404, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 28 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 25481 diverse cancer specimens. This rate is 1.64-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.66 % in 1119 large intestine cancers tested; 0.46 % in 575 stomach cancers tested; 0.34 % in 805 skin cancers tested; 0.31 % in 602 endometrium cancers tested; 0.25 % in 1943 lung cancers tested; 0.22 % in 605 oesophagus cancers tested; 0.16 % in 1270 liver cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,765 cancer specimens
Comments:
Only 2 deletions, 5 insertions and no complex mutations are noted on the COSMIC website.

