Nomenclature
Short Name:
ErbB3
Full Name:
Receptor tyrosine-protein kinase erbB-3
Alias:
- C-erbB3
- Receptor protein-tyrosine kinase erbB-3 precursor
- Tyrosine kinase-type cell surface receptor HER3
- V-erb-b2 erythroblastic leukemia viral oncogene 3
- EC 2.7.10.1
- ERB3
- HER3
- Kinase ErbB3
- LCCS2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
EGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
148,098
# Amino Acids:
1342
# mRNA Isoforms:
5
mRNA Isoforms:
148,098 Da (1342 AA; P21860); 141,752 Da (1283 AA; P21860-4); 77,426 Da (699 AA; P21860-5); 36,490 Da (331 AA; P21860-3); 20,136 Da (183 AA; P21860-2)
4D Structure:
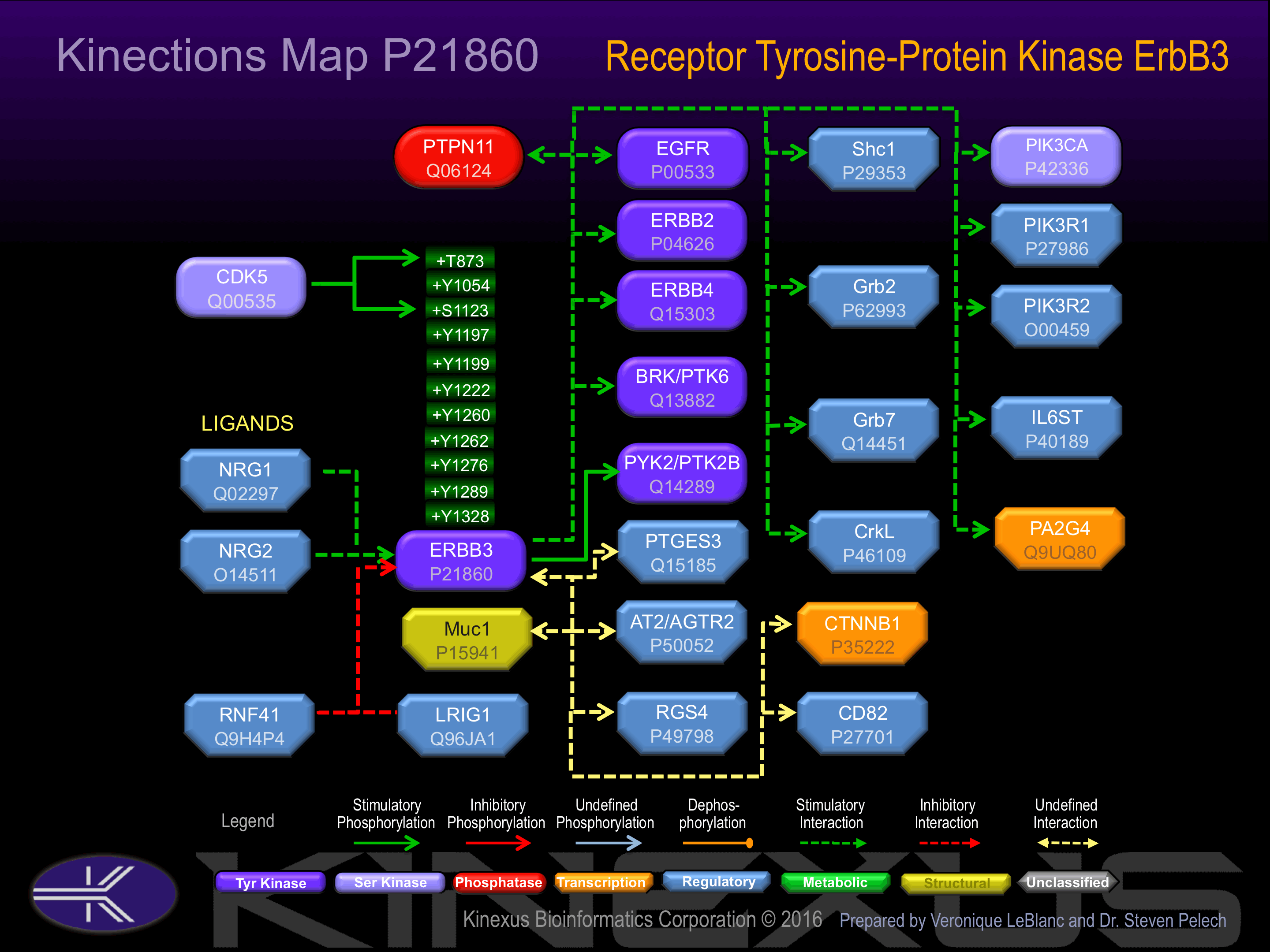
Heterodimer with each of the other ERBB receptors Potential. Interacts with CSPG5, PA2G4 and MUC1
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
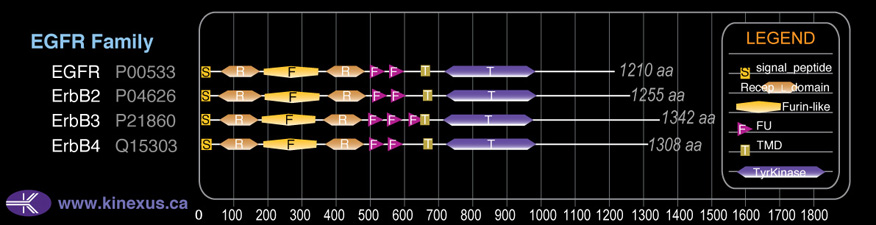
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 19 | signal_peptide |
| 55 | 167 | Recep_L_domain |
| 180 | 332 | Furin-like |
| 353 | 474 | Recep_L_domain |
| 490 | 541 | FU |
| 546 | 595 | FU |
| 607 | 642 | FU |
| 644 | 666 | TMD |
| 709 | 965 | TyrKc |
| 709 | 967 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
K736.
N-GlcNAcylated:
N126, N250, N353, N408, N414, N437, N469, N522, N566, N616.
Serine phosphorylated:
S686, S844, S846, S869, S982, S1123, S1147, S1207, S1215, S1285, S1315, S1331.
Threonine phosphorylated:
T640+, T873+, T1164, T1319.
Tyrosine phosphorylated:
Y680, Y868, Y1054, Y1132, Y1159, Y1197, Y1199, Y1222, Y1260, Y1262, Y1276, Y1289, Y1307, Y1328.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 52
52
1148
47
1428
 2
2
39
27
39
 4
4
87
29
83
 13
13
278
211
438
 52
52
1139
49
753
 5
5
119
101
176
 37
37
806
85
721
 99
99
2174
82
6470
 38
38
832
17
776
 16
16
347
180
434
 7
7
143
73
169
 20
20
438
356
605
 3
3
55
51
117
 2
2
34
27
37
 10
10
214
65
230
 0.8
0.8
18
34
14
 11
11
247
725
1714
 11
11
232
48
339
 2
2
52
165
44
 36
36
788
183
700
 9
9
196
66
255
 2
2
41
63
76
 9
9
204
47
145
 2
2
44
48
55
 3
3
59
64
160
 47
47
1044
136
2783
 4
4
94
54
91
 6
6
140
49
124
 3
3
58
46
67
 5
5
113
42
64
 19
19
410
36
289
 100
100
2200
57
5190
 34
34
739
107
1260
 32
32
704
140
666
 7
7
158
74
210
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.9
100 98.7
98.7
99.3
98 -
-
-
93 -
-
-
- 81.1
81.1
83.1
94 -
-
-
- 90.6
90.6
94.3
91 90.5
90.5
94.6
91 -
-
-
- 48.4
48.4
63.9
- 25.9
25.9
35.3
71 47
47
63.3
62 48.4
48.4
61.6
- -
-
-
- 25
25
39.8
- -
-
-
- 21.2
21.2
38.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PIK3R1 - P27986 |
| 2 | ERBB2 - P04626 |
| 3 | PA2G4 - Q9UQ80 |
| 4 | SHC1 - P29353 |
| 5 | RNF41 - Q9H4P4 |
| 6 | ERBB4 - Q15303 |
| 7 | EGFR - P00533 |
| 8 | NRG2 - O14511 |
| 9 | GRB7 - Q14451 |
| 10 | PTK6 - Q13882 |
| 11 | GRB2 - P62993 |
| 12 | PIK3R2 - O00459 |
| 13 | EGF - P01133 |
| 14 | PTK2B - Q14289 |
| 15 | AGTR2 - P50052 |
Regulation
Activation:
Binds and is activated by neuregulins and NTAK. Phosphorylation of Tyr-1222 and Tyr-1260 induces interaction with PIK3C and PIK3R1. Kinase domain lacks activity but heterodimerizes with other EGFRs to transduce growth signals.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
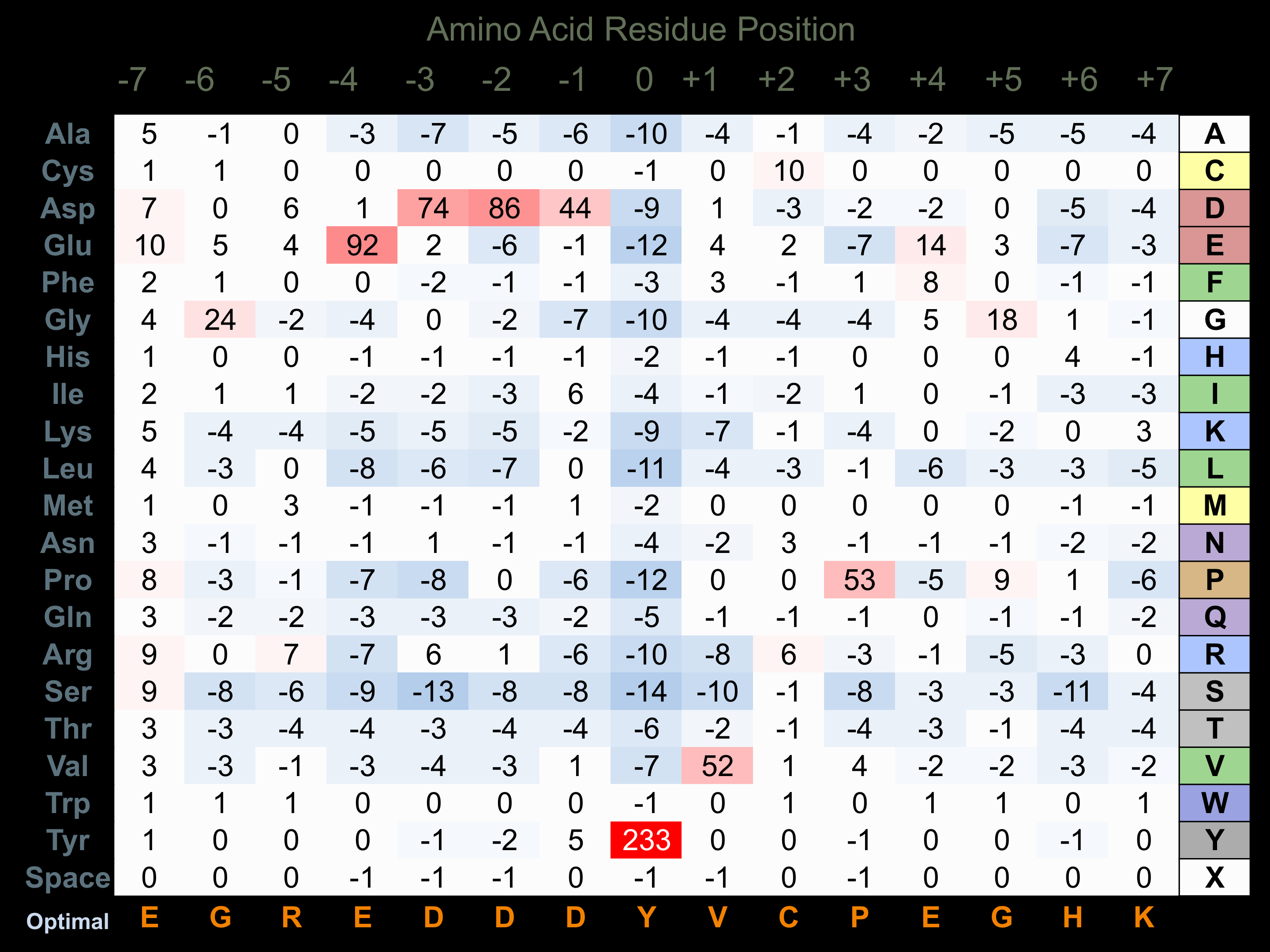
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Bosutinib | Kd = 770 pM | 5328940 | 288441 | 22037378 |
| AZD8931 | IC50 = 4 nM | 11488320 | ||
| Neratinib | Kd = 7.7 nM | 9915743 | 180022 | 22037378 |
| Dasatinib | Kd = 18 nM | 11153014 | 1421 | 22037378 |
| PP242 | Kd = 120 nM | 25243800 | 22037378 | |
| PD173955 | Kd = 140 nM | 447077 | 386051 | 22037378 |
| Vandetanib | Kd = 160 nM | 3081361 | 24828 | 22037378 |
| Cediranib | Kd = 180 nM | 9933475 | 491473 | 22037378 |
| Canertinib | Kd = 210 nM | 156414 | 31965 | 22037378 |
| Gefitinib | Kd = 790 nM | 123631 | 939 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Erlotinib | Kd = 1.1 µM | 176870 | 553 | 22037378 |
| Foretinib | Kd = 1.5 µM | 42642645 | 1230609 | 22037378 |
| GDC0879 | Kd = 2.3 µM | 11717001 | 525191 | 22037378 |
| Afatinib | Kd = 4.5 µM | 10184653 | 1173655 | 22037378 |
Disease Linkage
General Disease Association:
Cancer, neurological, and physical disorders
Specific Diseases (Non-cancerous):
Congenital contractures; Lethal congenital contracture syndrome 2; Charcot-Marie-Tooth disease Type 1a; Charcot-Marie-Tooth disease Type 1
Comments:
Congenital Contractures, a rare disease, is characterized with long limbs, long extremeties, permanently bent joints, underdeveloped muscles, and a protruding chest. Charcot-Marie-Tooth Disease Type 1a is a rare neuronal disease which results in weakness and wasting of lower limbs.
Specific Cancer Types:
Medulloblastomas; Breast cancer; Pancreatic cancer; Verrucous carcinomas; Childhood medulloblastomas; Carotid body tumours (CBT); Gestational trophoblastic tumours (GTD)
Comments:
ERBB3 might be an oncoprotein (OP) or a tumour suppressor protein (TSP). tumour suppressor protein (TSP)ErbB3 appears to be up-regulated in many cancers, although it is believed to be catalytically inactive as a protein kinase. ErbB3 is linked to Medulloblastomas, which is a rare, highly malignant brain tumour. Verrucous carcinoma is a squamous cell cancer that is non-metastatic. Childhood medulloblastoma is linked to primitive neuroectodermal tumour and can affect bone, brain, and thyroid tissues. Carotid body tumour (CBT) can affect thyroid, bone, and lymph node tissues and CBT is a rare neuroendocrine neoplasm. Gestational Trophoblastic tumour (GTD) is a pregnancy related tumour.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +319, p<0.003); Brain glioblastomas (%CFC= +6922, p<0.0001); Brain oligodendrogliomas (%CFC= +7016, p<0.0001); Cervical epithelial cancer (%CFC= +69, p<0.097); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +748, p<0.008); Colorectal adenocarcinomas (early onset) (%CFC= -53, p<0.014); Lung adenocarcinomas (%CFC= +72, p<0.0002); Ovary adenocarcinomas (%CFC= +78, p<0.04); Papillary thyroid carcinomas (PTC) (%CFC= +119, p<(0.0003); Prostate cancer - primary (%CFC= +538, p<0.0001); and Skin melanomas - malignant (%CFC= +270, p<0.0001). The COSMIC website notes an up-regulated expression score for ERBB3 in diverse human cancers of 438, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 28 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25734 diverse cancer specimens. This rate is only 14 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.56 % in 590 stomach cancers tested; 0.49 % in 152 biliary tract cancers tested; 0.42 % in 500 urinary tract cancers tested; 0.36 % in 1128 large intestine cancers tested; 0.2 % in 806 skin cancers tested; 0.15 % in 602 endometrium cancers tested; 0.09 % in 1498 breast cancers tested; 0.07 % in 1270 liver cancers tested; 0.06 % in 1226 kidney cancers tested; 0.05 % in 1944 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D279Y (10); R667H (5).
Comments:
Only 4 deletions, 1 insertion, and 1 complex mutation are noted on the COSMIC website.