Nomenclature
Short Name:
JNK1
Full Name:
Mitogen-activated protein kinase 8
Alias:
- C-Jun N-terminal kinase 1
- EC 2.7.11.24
- JNK1A2
- MAPK8
- MK08
- JNK21B1/2; Kinase JNK; PRKM8; SAPK1; Stress-activated protein kinase JNK1
- JNK
- JNK1-alpha-2
- JNK-46
- JUN N-terminal kinase
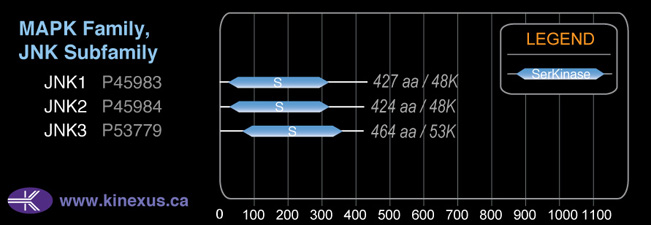
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
MAPK
SubFamily:
JNK
Specific Links
Structure
Mol. Mass (Da):
48,296
# Amino Acids:
427
# mRNA Isoforms:
5
mRNA Isoforms:
48,296 Da (427 AA; P45983); 48,088 Da (427 AA; P45983-4); 44,229 Da (384 AA; P45983-2); 44,022 Da (384 AA; P45983-3); 35,333 Da (308 AA; P45983-5)
4D Structure:
Binds to at least four scaffolding proteins, MAPK8IP1/JIP-1, MAPK8IP2/JIP-2, MAPK8IP3/JIP-3/JSAP1 and SPAG9/MAPK8IP4/JIP-4. These proteins also bind other components of the JNK signaling pathway. Interacts with TP53 and WWOX. Interacts with JAMP. Forms a
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 26 | 321 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K308.
Other:
C116 (S-nitrosocysteine).
Palmitoylated:
C425 (predicted), C426 (predicted).
Serine phosphorylated:
S129+, S155, S179, S292.
Threonine phosphorylated:
T178+, T183+, T188-, T243, T255, T258, T367.
Tyrosine phosphorylated:
Y185+, Y259, Y357.
Ubiquitinated:
K166, K300.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 44
44
1209
47
1005
 1.4
1.4
40
19
38
 4
4
113
12
119
 11
11
312
185
426
 29
29
798
55
590
 1.2
1.2
32
102
26
 7
7
189
69
378
 40
40
1118
67
2249
 10
10
275
17
230
 4
4
116
191
98
 2
2
58
46
55
 20
20
555
246
663
 1
1
28
34
29
 1.1
1.1
31
17
20
 6
6
168
40
668
 1.5
1.5
41
30
26
 5
5
128
567
1176
 21
21
571
22
2217
 4
4
99
153
76
 20
20
567
193
528
 5
5
131
39
227
 2
2
49
43
56
 2
2
61
22
56
 5
5
135
23
125
 4
4
118
39
128
 41
41
1140
118
1656
 2
2
47
37
67
 5
5
126
23
227
 3
3
83
23
117
 7
7
194
70
127
 17
17
458
24
451
 100
100
2772
57
7064
 25
25
680
109
1158
 31
31
862
135
745
 10
10
266
74
219
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 41
41
55.7
100 99.8
99.8
100
100 -
-
-
99 -
-
-
100 99.5
99.5
100
99.5 -
-
-
- 82.8
82.8
87.3
98 95.1
95.1
95.3
97 -
-
-
- 82.4
82.4
89.7
- 74.7
74.7
81.5
99 91.6
91.6
95.3
96 77.5
77.5
84.5
87 -
-
-
- 67
67
76.8
80 -
-
-
- 57
57
68
72 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAP2K4 - P45985 |
| 2 | ATF2 - P15336 |
| 3 | MAPK8IP1 - Q9UQF2 |
| 4 | IRS1 - P35568 |
| 5 | TP53 - P04637 |
| 6 | MAPK8IP3 - Q9UPT6 |
| 7 | GSTP1 - P09211 |
| 8 | DUSP4 - Q13115 |
| 9 | TNFSF11 - O14788 |
| 10 | PPARG - P37231 |
| 11 | BAD - Q92934 |
| 12 | NR3C1 - P04150 |
| 13 | JUND - P17535 |
| 14 | MAP1B - P46821 |
| 15 | JKAMP - Q9P055 |
Regulation
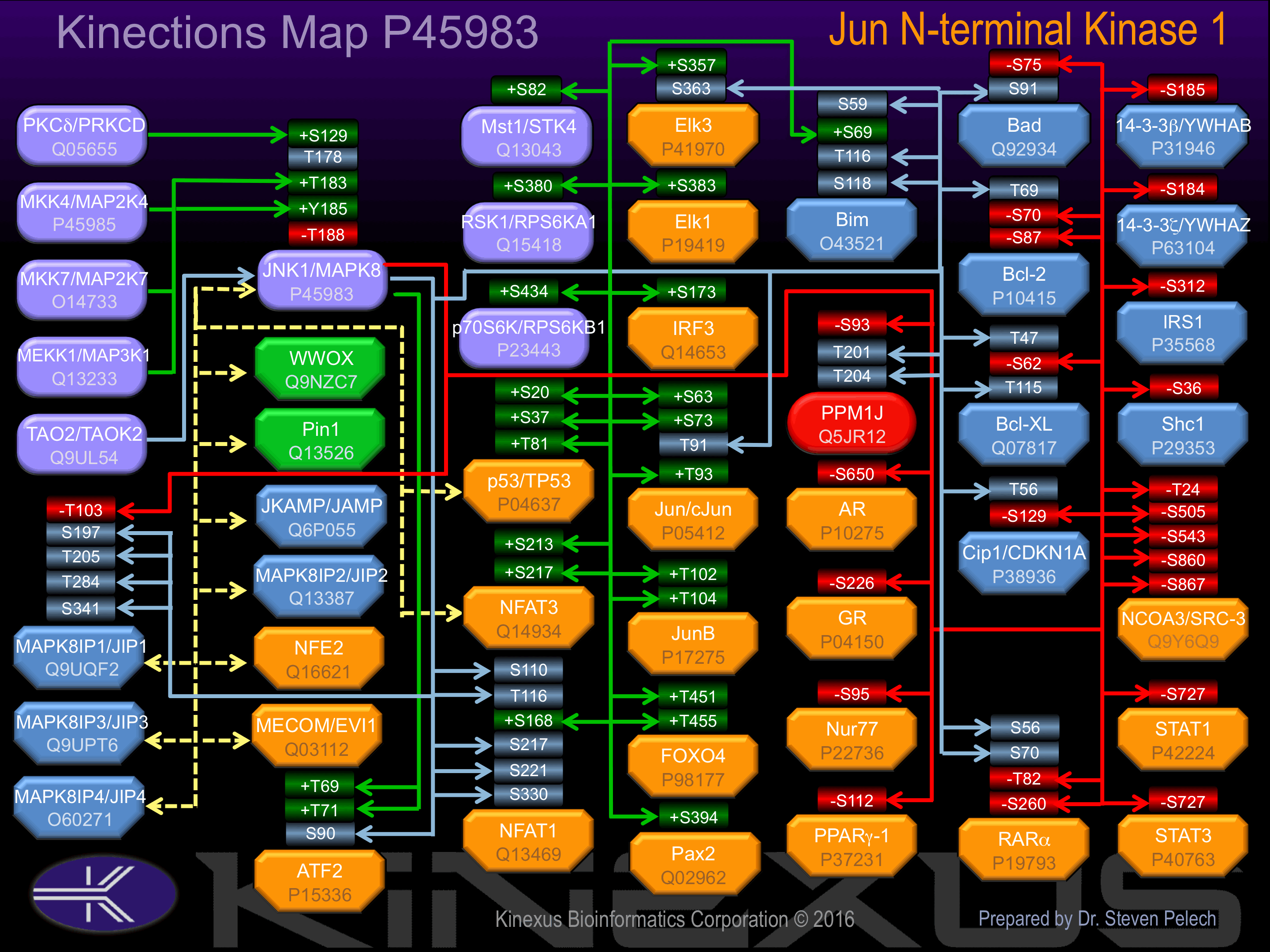
Activation:
Phosphorylation of Thr-183 and Tyr-185 by either of two dual specificity kinases, MAP2K4 and MAP2K7, increases phosphotransferase activity.
Inhibition:
Inhibited by dual specificity phosphatases, such as DUSP1, which target the Thr-183 and Tyr-185 activatory phosphorylation sites.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 14-3-3 beta (YWHAB) | P31946 | S185 | FYYEILNSPEKACSL | - |
| 14-3-3 sigma | P31947 | S186 | FHYEIANSPEEAISL | |
| 14-3-3 zeta (YWHAZ) | P63104 | S184 | FYYEILNSPEKACSL | - |
| APLP2 | Q06481 | T736 | VEVDPMLTPEERHLN | |
| APP | P05067 | T743 | VEVDAAVTPEERHLS | |
| AR | P10275 | S650 | GEASSTTSPTEETTQ | - |
| ATF2 | P15336 | S90 | GLFNELASPFENEFK | |
| ATF2 | P15336 | T69 | SVIVADQTPTPTRFL | + |
| ATF2 | P15336 | T71 | IVADQTPTPTRFLKN | + |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| Bad | Q92934 | S91 | EGMGEEPSPFRGRSR | |
| Bcl-2 | P10415 | S70 | RDPVARTSPLQTPAA | |
| Bcl-2 | P10415 | S87 | AAAGPALSPVPPVVH | + |
| Bcl-2 | P10415 | T69 | SRDPVARTSPLQTPA | |
| Bcl-xL | Q07817 | S62 | PSWHLADSPAVNGAT | |
| Bcl-xL | Q07817 | T115 | LTSQLHITPGTAYQS | |
| Bcl-xL | Q07817 | T47 | GTESEMETPSAINGN | |
| Bim | O43521 | S118 | DKSTQTPSPPCQAFN | |
| Bim | O43521 | S59 | GDSCPHGSPQGPLAP | |
| Bim | O43521 | S69 | GPLAPPASPGPFATR | |
| Bim | O43521 | T116 | SCDKSTQTPSPPCQA | |
| Bim iso2 | O43521-2 | S44 | QTEPQDRSPAPMSCD | |
| Bim iso2 | O43521-2 | S58 | DKSTQTPSPPCQAFN | |
| Bim iso2 | O43521-2 | T56 | SCDKSTQTPSPPCQA | |
| Cip1 (p21, CDKN1A) | P38936 | S129 | SGEQAEGSPGGPGDS | - |
| Cip1 (p21, CDKN1A) | P38936 | T56 | NFDFVTETPLEGDFA | |
| Ctbp1 | Q13363 | S422 | AHPPHAPSPGQTVKP | |
| CTNNB1 | P35222 | S191 | SRHAIMRSPQMVSAI | |
| CTNNB1 | P35222 | S605 | LFVQLLYSPIENIQR | |
| DLG4 (PSD-95) | P78352 | S295 | PTSPRRYSPVAKDLL | |
| Doublecortin | O43602 | S415 | SPISTPTSPGSLRKH | |
| Doublecortin | O43602 | T402 | TSSSQLSTPKSKQSP | |
| Doublecortin | O43602 | T412 | SKQSPISTPTSPGSL | |
| DRPLA | P54259 | S734 | EEYETPESPVPPARS | |
| Elk1 | P19419 | S383 | IHFWSTLSPIAPRSP | + |
| Elk1 | P19419 | S389 | LSPIAPRSPAKLSFQ | |
| Elk3 | P41970 | S357 | IHFWSSLSPVAPLSP | |
| Elk3 | P41970 | S363 | LSPVAPLSPARLQGP | |
| FOXO4 | P98177 | T451 | PIPKALGTPVLTPPT | + |
| FOXO4 | P98177 | T455 | ALGTPVLTPPTEAAS | + |
| GR | P04150 | S226 | IDENCLLSPLAGEDD | - |
| H3.2 | P84228 | S29 | ATKAARKSAPATGGV | |
| hnRNP K | P61978 | S216 | ILDLISESPIKGRAQ | + |
| hnRNP K | P61978 | S353 | DSAIDTWSPSEWQMA | |
| HSF1 | Q00613 | S363 | DTEGRPPSPPPTSTP | |
| IRF3 | Q14653 | S173 | PCPQPLRSPSLDNPT | + |
| IRS1 | P35568 | S312 | TESITATSPASMVGG | - |
| IRS2 | Q9Y4H2 | S491 | GSASASGSPSDPGFM | |
| IRS2 | Q9Y4H2 | T350 | RTDSLAATPPAAKCS | |
| ITCH | Q96J02 | S240 | RRVSGNNSPSLSNGG | |
| ITCH | Q96J02 | S273 | RPASVNGSPSATSES | |
| ITCH | Q96J02 | T263 | PSRPPPPTPRRPASV | |
| JDP2 | Q8WYK2 | T148 | VRTDSVKTPESEGNP | |
| JIP1 | Q9UQF2 | S197 | DRVSRSSSPLKTGEQ | |
| JIP1 | Q9UQF2 | S341 | EGFDCLSSPERAEPP | |
| JIP1 | Q9UQF2 | T103 | LIDATGDTPGAEDDE | - |
| JIP1 | Q9UQF2 | T205 | PLKTGEQTPPHEHIC | |
| JIP1 | Q9UQF2 | T284 | ATEEIYLTPVQRPPD | |
| Jun (c-Jun) | P05412 | S63 | KNSDLLTSPDVGLLK | + |
| Jun (c-Jun) | P05412 | S73 | VGLLKLASPELERLI | + |
| Jun (c-Jun) | P05412 | T91 | SNGHITTTPTPTQFL | |
| Jun (c-Jun) | P05412 | T93 | GHITTTPTPTQFLCP | + |
| JunB | P17275 | T102 | SNGVITTTPTPPGQY | |
| JunB | P17275 | T104 | GVITTTPTPPGQYFY | |
| JunD | P17535 | S100 | LGLLKLASPELERLI | + |
| JunD | P17535 | S90 | PPDGLLASPDLGLLK | + |
| JunD | P17535 | T117 | SNGLVTTTPTSSQFL | |
| KRT8 | P05787 | S74 | TVNQSLLSPLVLEVD | |
| LAT | O43561 | T184 | PSAPALSTPGIRDSA | |
| LAT iso2 | O43561 | T155 | PSAPALSTPGIRDSA | |
| MCL1 | Q07820 | S121 | PAADAIMSPEEELDG | |
| MCL1 | Q07820 | S64 | IGGSAGASPPSTLTP | |
| MCL1 | Q07820 | T163 | TDGSLPSTPPPAEEE | |
| MST1 (STK4) | Q13043 | S82 | SIMQQCDSPHVVKYY | + |
| Myc | P01106 | S62 | LLPTPPLSPSRRSGL | |
| Myc | P01106 | S71 | SRRSGLCSPSYVAVT | |
| NCOA3 (SRC-3) | Q9Y6Q9 | S505 | SPVAGVHSPMASSGN | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S543 | SLLSTLSSPGPKLDN | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S860 | NRAVSLDSPVSVGSS | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S867 | SPVSVGSSPPVKNIS | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | T24 | KRKLPCDTPGQGLTC | - |
| NFAT1 | Q13469 | S110 | PAGASGLSPRIEITP | |
| NFAT1 | Q13469 | S168 | YREPLCLSPASSGSS | |
| NFAT1 | Q13469 | S217 | AHYSPRTSPIMSPRT | |
| NFAT1 | Q13469 | S221 | PRTSPIMSPRTSLAE | |
| NFAT1 | Q13469 | S330 | WKTSPDPSPVSAAPS | |
| NFAT1 | Q13469 | T116 | LSPRIEITPSHELIQ | |
| NFAT3 | Q14934 | S213 | ASRFGLGSPLPSPRA | + |
| NFAT3 | Q14934 | S217 | GLGSPLPSPRASPRP | + |
| Nur77 | P22736 | S95 | TSSSSATSPASASFK | - |
| p53 (TP53) | P04637 | S20 | PLSQETFSDLWKLLP | + |
| p53 (TP53) | P04637 | S37 | NVLSPLPSQAMDDLM | + |
| p53 (TP53) | P04637 | T81 | APAPAAPTPAAPAPA | ? |
| p70S6K (RPS6KB1) | P23443 | S434 | SFEPKIRSPRRFIGS | + |
| Pax2 | Q02962 | S394 | SNPALLSSPYYYSAA | + |
| Pax2 | Q02962 | T297 | EYSLPALTPGLDEVK | |
| PPARg-1 | P37231 | S112 | AIKVEPASPPYYSEK | - |
| PPM1J | Q5JR12 | S93 | HAGRAVQSPPDTGRR | - |
| PPM1J | Q5JR12 | T201 | PPLCLPTTPGTPDSS | |
| PPM1J | Q5JR12 | T204 | CLPTTPGTPDSSDPS | |
| PXN iso2 | P49023 | S178 | PPLPGALSPLYGVPE | |
| RSK1 (RPS6KA1) | Q15418 | S380 | HQLFRGFSFVATGLM | + |
| RXRa | P19793 | S260 | NMGLNPSSPNDPVTN | - |
| RXRa | P19793 | S32 | RGSMAAPSLHPSLGP | |
| RXRa | P19793 | S56 | SPISTLSSPINGMGP | |
| RXRa | P19793 | S70 | PPFSVISSPMGPHSM | |
| RXRa | P19793 | T82 | HSMSVPTTPTLGFST | - |
| Shc1 | P29353 | S36 | TPPEELPSPSASSLG | - |
| SP1 | P08047 | T278 | SVSAATLTPSSQAVT | |
| SP1 | P08047 | T739 | SEGSGTATPSALITT | |
| SPIB | Q01892 | T56 | VAPPVPATPYEAFDP | |
| STAT1 | P42224 | S727 | TDNLLPMSPEEFDEM | - |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| STMN2 | Q93045 | S62 | ELILKPPSPISEAPR | |
| STMN2 | Q93045 | S73 | EAPRTLASPKKKDLS | |
| Tau iso5 (Tau-C) | P10636-5 | S202 | SGYSSPGSPGTPGSR | |
| Tau iso5 (Tau-C) | P10636-5 | T205 | SSPGSPGTPGSRRTP | |
| Tau iso8 | P10636-8 | S396 | GAEIVYKSPVVSGDT | |
| Tau iso8 | P10636-8 | T181 | KTPPAPKTPPSSGEP | |
| Tau iso8 | P10636-8 | T231 | KKVAVVRTPPKSPSS | |
| Tau iso9 (Tau-F) | P10636-9 | S404 | PVVSGDTSPRHLSNV | |
| TIF-IA | Q9NYV6 | T200 | IARYVPSTPWFLMPI | |
| WWOX | Q9NZC7 | Y33 | TTKDGWVYYANHTEE |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
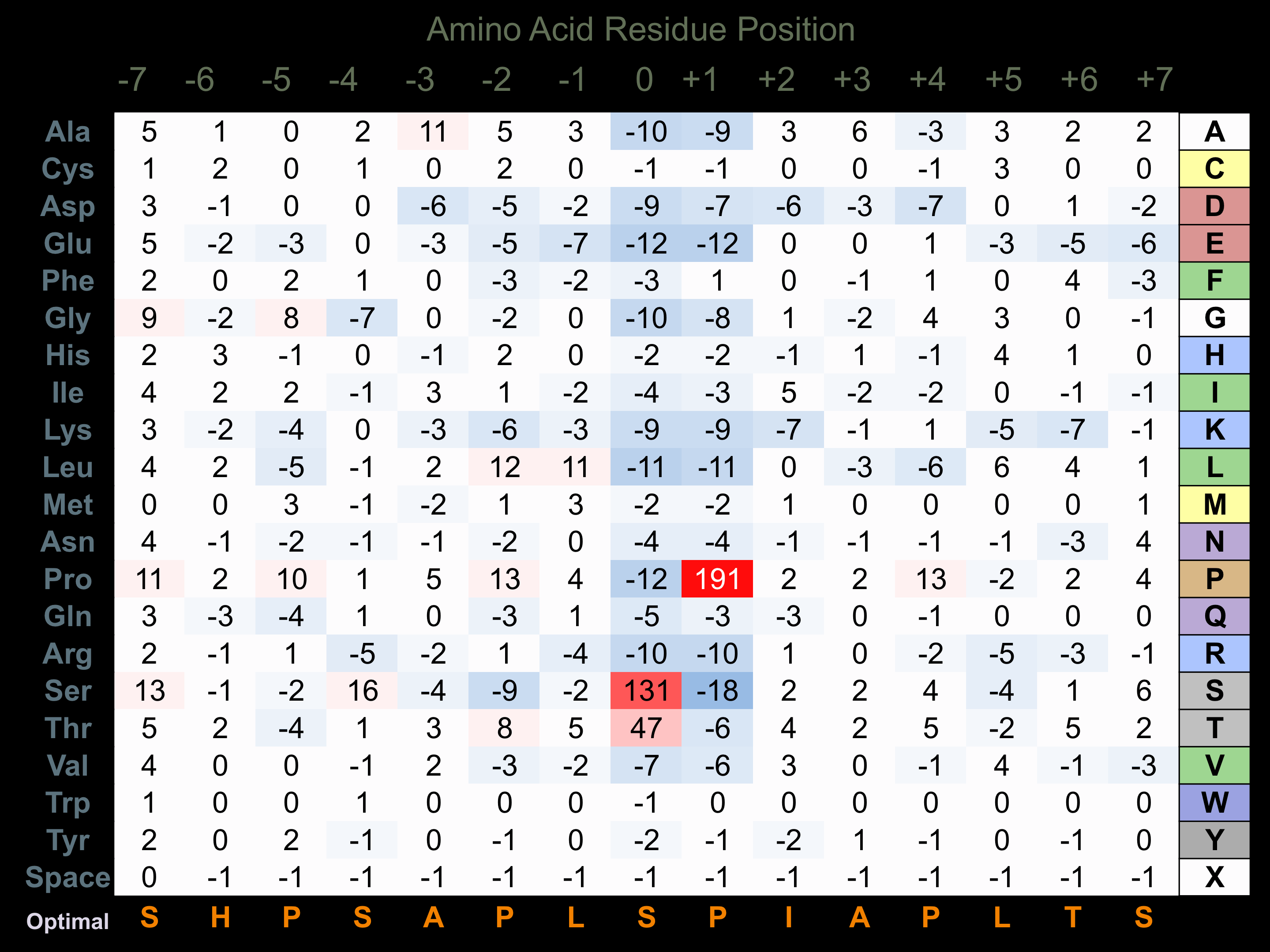
Matrix Type:
Experimentally derived from alignment of 162 known protein substrate phosphosites and 11 peptides phosphorylated by recombinant JNK1 in vitro tested in-house by Kinexus. Note that additional binding sites on JNK substrates with D motifs (consensus=R-P-t-s/r/t-L-p-L or K/r-x-s-L-s-L/i/v-s-l/p) facilitate higher selectivity for phosphorylation by this protein kinase.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cardiovascular disorders
Specific Diseases (Non-cancerous):
Diastolic heart failure; Restrictive cardiomyopathy; Frozen shoulder
Comments:
Diastolic heart failure is a cardiovascular disease characterized by the funtional decline in either the left ventricle or both ventricles during diastole. Diastolic heart failure can be caused by any process that leads to the stiffening fo the left ventricle, including hypertension, aortic stenosis, diabetes, constrictive pericarditis, restrictive cardiomyopathy. Restrictive cardiomyopathy is a heart disease characterized by rigid walls of the heart, restricting the stretching of the heart and thus the ability of the heart chamber to completely fill with blood. Frozen shoulder, also known as adhesive capsulitis, is a disease characterized by the inflammation of the connective tissue surrounding the shoulder, resulting in the restriction of movement of the glenohumeral joint of the shoulder. High levels of JNK phosphorylation have been observed in tissue samples from abdominal aortic aneurysm patients. In rat aortic vascular smooth muscle cells, JNK induces the expression of genes that promote the degradation of the extracellular matrix (ECM), while suppressing enzymes that synthesize ECM components, such as Lox and Plod1. Therefore, JNK is involved in the pathogenesis of disorders of the cardiovascular system as it acts to promote abnormal ECM metabolism. Activation of the stress-activated MAP kinases, particularly JNK and p38, are associated with different forms of cardiac pathology, specifically related to the role of the proteins in the regulation of hypertrophy and apoptosis. Increased JNK activity has been correlated with heart failure, specifically at the onset of stress stimulation. in this context, JNK may function to promote cardiomyocyte hypertrophy and apoptotic cell death. In addition, JNK activity has been linked to different aspects of cardiac pathology, including contractile dysfunction, extracellular matrix remodelling, communication between cardiomyocytes, and cardiac metabolic regulation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -52, p<0.0001); Brain glioblastomas (%CFC= +113, p<0.1); Cervical cancer stage 1B (%CFC= +385); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +384, p<0.0001); andProstate cancer - primary (%CFC= +48, p<0.0001). The COSMIC website notes an up-regulated expression score for JNK1 in diverse human cancers of 328, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 152 for this protein kinase in human cancers was 2.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25587 diverse cancer specimens. This rate is only -2 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.54 % in 1093 large intestine cancers tested; 0.43 % in 602 endometrium cancers tested; 0.32 % in 805 skin cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,845 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.