Nomenclature
Short Name:
MEKK1
Full Name:
Mitogen-activated protein kinase kinase kinase 1 [Fragment]
Alias:
- EC 2.7.11.25
- Kinase MEKK1
- MEK kinase 1
- MEKK
- MEKK1
- Mitogen-activated protein kinase kinase kinase 1
- M3K1
- MAP3K1
- MAPK/ERK kinase kinase 1
- MAPKKK1
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE11
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
164470
# Amino Acids:
1512
# mRNA Isoforms:
1
mRNA Isoforms:
164,470 Da (1512 AA; Q13233)
4D Structure:
Binds both upstream activators and downstream substrates in multimolecular complexes through its N-terminus. Oligomerizes after binding MAP4K2 or TRAF2. Interacts with AXIN1. Interacts (via the kinase catalytic domain) with STK38
1D Structure:
Subfamily Alignment

Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S21, S35, S67-, S133, S137, S154, S232, S242, S250, S252, S264, S266, S275, S281, S292, S297, S300, S333, S429, S430, S431, S507, S511, S531, S796, S844, S923, S925, S926, S1016, S1018, S1026, S1028, S1043, S1087, S1089, S1157, S1159, S1455, S1468, S1477, S1480.
Sumoylated:
K436.
Threonine phosphorylated:
T20, T285, T522, T542, T657, T665, T850, T920, T999, T1082, T1114+, T1400+, T1402, T1412-, T1414, T1471.
Tyrosine phosphorylated:
Y1338.
Ubiquitinated:
K690, K1168, K1176, K1398.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 40
40
1328
25
1587
 0.7
0.7
23
10
38
 0.1
0.1
3
1
0
 18
18
612
76
1743
 51
51
1697
18
728
 51
51
1705
54
4216
 10
10
326
30
399
 62
62
2081
29
4244
 18
18
605
7
408
 5
5
183
81
215
 0.6
0.6
19
15
28
 12
12
404
122
536
 1.3
1.3
43
12
17
 0.5
0.5
18
7
8
 1
1
33
12
42
 1.3
1.3
44
13
52
 1.2
1.2
39
263
193
 0.6
0.6
19
7
8
 1.5
1.5
49
64
67
 28
28
925
81
786
 0.7
0.7
24
12
32
 3
3
103
14
184
 0.7
0.7
23
2
28
 0.6
0.6
20
7
27
 1.5
1.5
49
12
86
 24
24
790
48
820
 1.2
1.2
39
12
17
 0.4
0.4
12
6
6
 0.2
0.2
8
6
7
 2
2
77
28
72
 5
5
155
6
9
 100
100
3356
22
9195
 15
15
503
66
1622
 29
29
970
52
787
 6
6
210
35
228
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
99 -
-
-
98 -
-
-
94 -
-
-
88 91.9
91.9
94.1
94 -
-
-
- 89.9
89.9
92.9
90.5 89.2
89.2
92.1
90 -
-
-
- 77.3
77.3
82.1
- 77
77
81.4
83 -
-
-
78 40.3
40.3
45.8
74 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 33.8
33.8
50.3
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by phosphorylation at Thr-1400 and Thr-1412. Phosphorylation of Ser-67 inhibits interaction with JNK1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| FADD | Q13158 | S194 | QNRSGAMSPMSWNSD | |
| IKKb (IKBKINASE) | O14920 | S177 | AKELDQGSLCTSFVG | + |
| JNK1 (MAPK8) | P45983 | T183 | AGTSFMMTPYVVTRY | + |
| JNK1 (MAPK8) | P45983 | Y185 | TSFMMTPYVVTRYYR | + |
| MEK1 (MAP2K1) | Q02750 | S218 | VSGQLIDSMANSFVG | + |
| MEK1 (MAP2K1) | Q02750 | S222 | LIDSMANSFVGTRSY | + |
| MEK1 (MAP2K1) | Q02750 | T292 | ETPPRPRTPGRPLSS | + |
| MEKK1 (MAP3K1) | Q13233 | T1400 | ARLASKGTGAGEFQG | + |
| MEKK1 (MAP3K1) | Q13233 | T1412 | FQGQLLGTIAFMAPE | - |
| MKK4 (MAP2K4, MEK4) | P45985 | S257 | ISGQLVDSIAKTRDA | + |
| MKK4 (MAP2K4, MEK4) | P45985 | T261 | LVDSIAKTRDAGCRP | + |
| MKK6 (MAP2K6, MEK6) | P52564 | S207 | ISGYLVDSVAKTIDA | + |
| MKK6 (MAP2K6, MEK6) | P52564 | T211 | LVDSVAKTIDAGCKP | + |
| MKK7 (MAP2K7, MEK7) | O14733 | S271 | ISGRLVDSKAKTRSA | + |
| MKK7 (MAP2K7, MEK7) | O14733 | T275 | LVDSKAKTRSAGCAA | + |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
| STAT3 | P40763 | Y705 | DPGSAAPYLKTKFIC | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
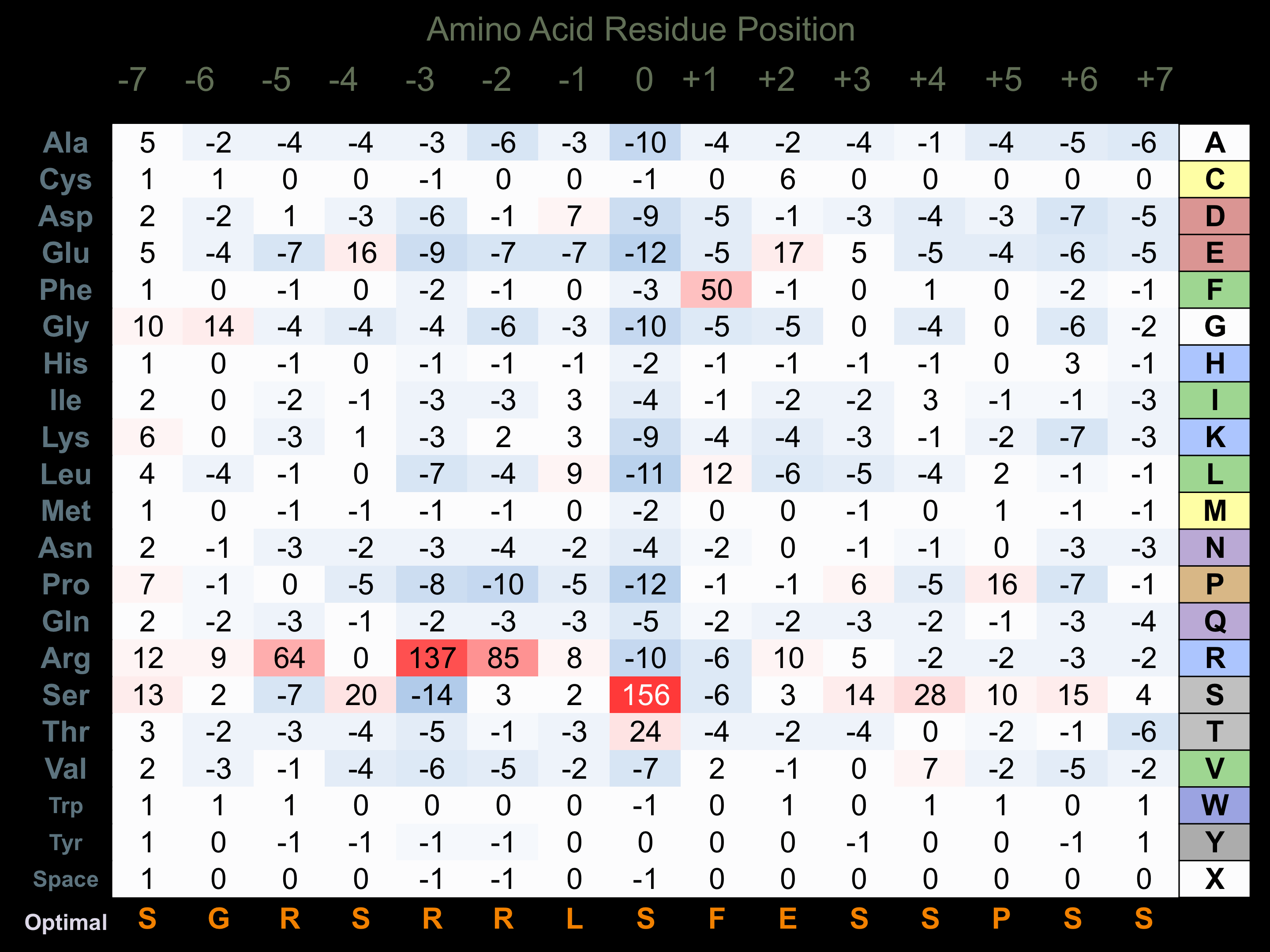
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, endocrine disorders
Specific Diseases (Non-cancerous):
46xy sex Reversal 6 (SRXY6); 46,xy disorder of sex development and 46,xy Complete gonadal dysgenesis; 46xy sex Reversal 3
Comments:
46xy Sex Reversal 6 (SRXY6) is a rare condition with symptoms including late puberty and polycystic ovaries. Affected tissues can include ovary, testes, and skin. A mutation in L189P can lead to binding of RHOA to MEKK1, and increased phosphorylation of MAPK3/MAPK1 which are downstream targets of MEKK1. The L189R mutation will lead to increased binding of RHOA to MEKK1, and increase phosphorylation of MAPK3/MAPK1 and of MAPK14. 46,xy Disorder of Sex Development and 46,xy Complete Gonadal Dysgenesis is a disorder linked to hermaphroditism, and gonadal dysgenesis. The rare condition 46xy Sex Reversal 3 can include symptoms such as gonadal dysgenesis, testis abnormalities, and testicular cancer.
Specific Cancer Types:
Breast cancer susceptibility; 46xy Sex Reversal 3
Comments:
MEKK1 may be a tumour suppressor protein in view of the higher frequency of deletions and insertions of the gene encoding this protein kinase in human tumours. Defects in MEKK1 have been linked with the rare condition 46xy Sex Reversal 3, which can include symptoms such as gonadal dysgenesis, testis abnormalities, and testicular cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Classical Hodgkin lymphomas (%CFC= -74, p<0.09); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +2168, p<0.0001); Large B-cell lymphomas (%CFC= -55, p<0.093); and Prostate cancer - primary (%CFC= +142, p<0.0001).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24440 diverse cancer specimens. This rate is only -6 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.23 % in 805 skin cancers tested; 0.23 % in 1093 large intestine cancers tested; 0.15 % in 500 urinary tract cancers tested; 0.14 % in 602 endometrium cancers tested; 0.11 % in 589 stomach cancers tested; 0.07 % in 1808 lung cancers tested; 0.07 % in 1292 breast cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,723 cancer specimens
Comments:
Over 50 deletions (19 at T949delT), and 29 insertions, and no complex mutations are noted on the COSMIC website.

