Nomenclature
Short Name:
MEK7
Full Name:
Dual specificity mitogen-activated protein kinase kinase 7
Alias:
- C-Jun N-terminal kinase kinase 2
- EC 2.7.12.2
- Jun N-terminal kinase kinase 2
- MAP2K7
- MKK7
- PRKMK7
- JNK activating kinase 2
- JNK kinase 2
- JNK-activating kinase 2
- JNKK2
Classification
Type:
Dual specificity protein kinase
Group:
STE
Family:
STE7
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
47,485
# Amino Acids:
419
# mRNA Isoforms:
4
mRNA Isoforms:
51,881 Da (462 AA; O14733-2); 49,071 Da (435 AA; O14733-3); 48,182 Da (426 AA; O14733-4); 47,485 Da (419 AA; O14733)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 30 | Coiled-coil |
| 120 | 380 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S35, S61, S79, S86, S89, S271+, S277, S378, S403, S407, S411.
Threonine phosphorylated:
T66, T83, T142, T275+, T401, T406.
Tyrosine phosphorylated:
Y283.
Ubiquitinated:
K95, K245, K373.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1134
51
1180
 3
3
31
22
29
 16
16
179
13
376
 35
35
402
177
1067
 57
57
651
53
561
 12
12
140
136
527
 8
8
90
63
265
 56
56
640
56
970
 28
28
320
24
260
 6
6
67
160
84
 7
7
75
39
124
 56
56
632
224
529
 3
3
39
46
61
 2
2
28
15
26
 12
12
136
31
228
 5
5
52
32
60
 7
7
75
216
87
 15
15
167
24
508
 8
8
91
157
115
 41
41
467
218
480
 8
8
93
28
165
 7
7
82
34
170
 6
6
69
16
66
 8
8
95
24
218
 11
11
130
28
236
 53
53
600
111
538
 4
4
50
49
108
 7
7
80
23
168
 5
5
56
24
58
 31
31
346
70
370
 42
42
472
30
574
 90
90
1025
61
2544
 7
7
83
113
330
 66
66
749
130
650
 7
7
77
83
94
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 77
77
78.2
100 83.5
83.5
83.8
- -
-
-
99.5 -
-
-
- 99
99
99.5
99 -
-
-
- 77.2
77.2
77.6
99 99
99
99.5
99 -
-
-
- -
-
-
- -
-
-
- 40.4
40.4
54.5
86 39.4
39.4
56.8
82.5 -
-
-
- 22.4
22.4
27.6
63 35.3
35.3
43.3
- 34.8
34.8
52
54 43.7
43.7
54.4
- -
-
-
- -
-
-
- -
-
-
- 29.6
29.6
48.2
- 25.6
25.6
39.7
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
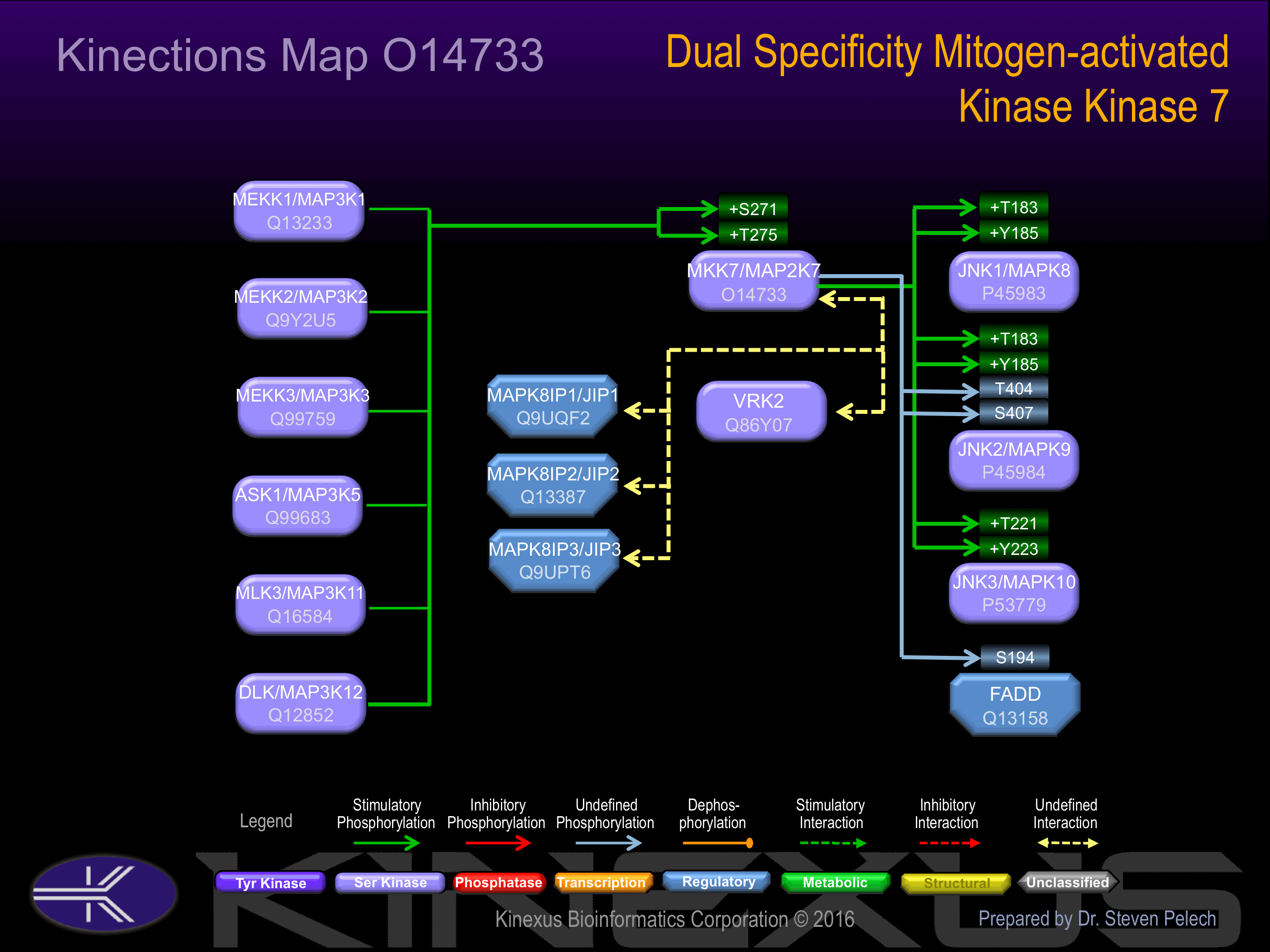
Activated by phosphorylation at Ser-271 and Thr-275 in the kinase activation loop by specific MAP kinase kinase kinases including MAP3K1 (MEKK1), MAP3K3 (MEKK3), MAP3K11 (MLK3) and MAP3K12 (DLK).
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ASK1 | Q99683 | S271 | ISGRLVDSKAKTRSA | + |
| MEKK1 | Q13233 | S271 | ISGRLVDSKAKTRSA | + |
| MEKK2 | Q9Y2U5 | S271 | ISGRLVDSKAKTRSA | + |
| MEKK3 | Q99759 | S271 | ISGRLVDSKAKTRSA | + |
| MLK3 | Q16584 | S271 | ISGRLVDSKAKTRSA | + |
| DLK | Q12852 | S271 | ISGRLVDSKAKTRSA | + |
| ASK1 | Q99683 | T275 | LVDSKAKTRSAGCAA | + |
| MEKK1 | Q13233 | T275 | LVDSKAKTRSAGCAA | + |
| MEKK2 | Q9Y2U5 | T275 | LVDSKAKTRSAGCAA | + |
| MEKK3 | Q99759 | T275 | LVDSKAKTRSAGCAA | + |
| MLK3 | Q16584 | T275 | LVDSKAKTRSAGCAA | + |
| DLK | Q12852 | T275 | LVDSKAKTRSAGCAA | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| FADD | Q13158 | S194 | QNRSGAMSPMSWNSD | |
| JNK1 (MAPK8) | P45983 | T183 | AGTSFMMTPYVVTRY | + |
| JNK1 (MAPK8) | P45983 | Y185 | TSFMMTPYVVTRYYR | + |
| JNK2 (MAPK9) | P45984 | S407 | STEQTLASDTDSSLD | |
| JNK2 (MAPK9) | P45984 | T183 | ACTNFMMTPYVVTRY | + |
| JNK2 (MAPK9) | P45984 | T404 | SSMSTEQTLASDTDS | |
| JNK2 (MAPK9) | P45984 | Y185 | TNFMMTPYVVTRYYR | + |
| JNK3 (MAPK10) | P53779 | T221 | AGTSFMMTPYVVTRY | + |
| JNK3 (MAPK10) | P53779 | Y223 | TSFMMTPYVVTRYYR | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
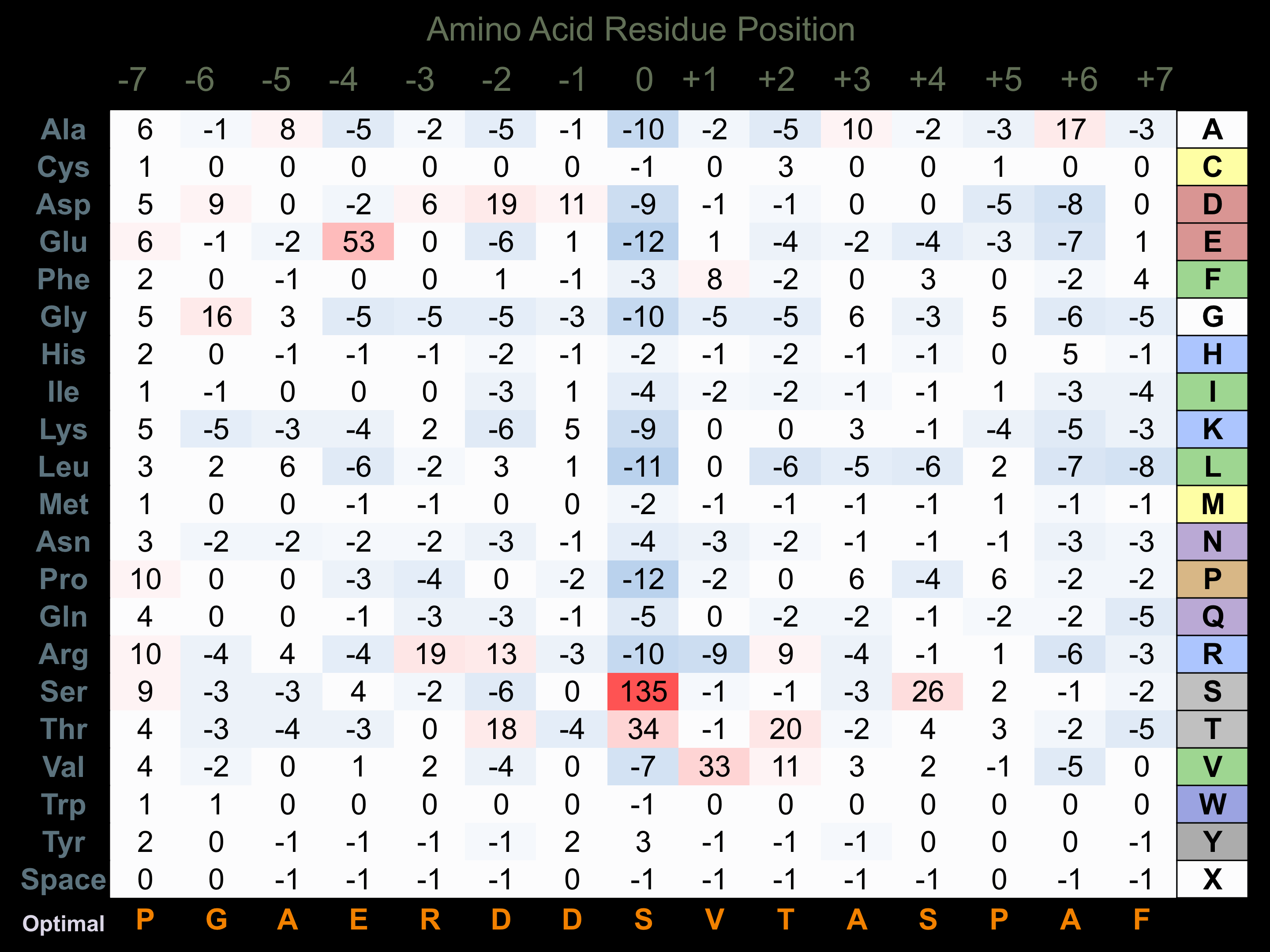
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AST-487 | Kd = 27 nM | 11409972 | 574738 | 22037378 |
| Staurosporine | Kd = 30 nM | 5279 | 22037378 | |
| Lestaurtinib | Kd = 80 nM | 126565 | 22037378 | |
| AT9283 | IC50 > 100 nM | 24905142 | 19143567 | |
| Canertinib | Kd = 110 nM | 156414 | 31965 | 22037378 |
| Neratinib | Kd = 530 nM | 9915743 | 180022 | 22037378 |
| CHEMBL215803 | IC50 > 1 µM | 16046126 | 215803 | 16876403 |
| CHEMBL307152 | IC50 < 1 µM | 11655119 | 307152 | 12824014 |
| CP673451 | IC50 > 1 µM | 10158940 | 15705896 | |
| MK5108 | IC50 > 1 µM | 24748204 | 20053775 | |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| SNS314 | IC50 > 1 µM | 16047143 | 514582 | 18678489 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| CEP5104 | IC50 > 3 µM | 23378546 | 460989 | 18714982 |
| CEP6331 | IC50 > 3 µM | 9823787 | 460990 | 18714982 |
| JNJ-28871063 | IC50 > 4 µM | 17747413 | 17975007 | |
| Lapatinib | Kd = 4.4 µM | 208908 | 554 | 22037378 |
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Schizophrenia
Comments:
The development of schizophrenia has been linked to abnormal glutaminergic signalling in certain brain regions, particularly the prefrontal cortex (PFC). in addition, MEK7 activity has been shown to be stimulated by glutaminergic signalling in brain tissue. Significantly reduced expression of MEK7 has been reported in post-mortem brain tissue from patients with schizophrenia, indicating a role for the protein in the pathology of the disease. While no mutations in MEK7 have been observed in schizophrenia patients, certain MEK7 haplotypes associated with the disease have been shown to significantly influence expression levels of MEK7 mRNA in the human prefrontal cortex. The role of MEK7 in the pathogenesis of schizophrenia may be through altered expression levels rather than mutation of the protein. In animal studies, mice lacking one copy of the MEK7 gene displayed a cognitive phenotype resembling human schizophrenia, including the impairment of working memory. Therefore, reduced expression of MEK7 has been suggested to play an important role in the pathogenesis of schizophrenia.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -49, p<0.045); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +706, p<0.0005); Malignant pleural mesotheliomas (MPM) tumours (%CFC= -46, p<0.055); and Prostate cancer - primary (%CFC= +70, p<0.0001). The COSMIC website notes an up-regulated expression score for MEK7 in diverse human cancers of 500, which is 1.1-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 271 for this protein kinase in human cancers was 4.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.13 % in 25435 diverse cancer specimens. This rate is 1.7-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.22 % in 1119 large intestine cancers tested; 0.65 % in 589 stomach cancers tested; 0.42 % in 805 skin cancers tested; 0.12 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D243N (7); R153H (6).
Comments:
Only 4 deletions, 4 insertions, and no complex mutations are noted on the COSMIC website.