Nomenclature
Short Name:
MEK6
Full Name:
Dual specificity mitogen-activated protein kinase kinase 6
Alias:
- EC 2.7.12.2
- Kinase MKK6
- MKK6
- Mitogen-activated protein kinase kinase 6
- MP2K6
- SAPKK3
- MAP kinase kinase 6
- MAPK/ERK kinase 6
- MAP2K6
- MAPKK6
Classification
Type:
Dual specificity protein kinase
Group:
STE
Family:
STE7
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
37,492
# Amino Acids:
334
# mRNA Isoforms:
2
mRNA Isoforms:
37,492 Da (334 AA; P52564); 31,339 Da (278 AA; P52564-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 53 | 314 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K5, K8, K82, S207, T211.
Serine phosphorylated:
S27, S90, S201, S207+.
Threonine phosphorylated:
T28, T211+.
Tyrosine phosphorylated:
Y64, Y203, Y219-.
Ubiquitinated:
K172.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 46
46
1120
26
1462
 2
2
52
15
70
 4
4
98
13
73
 9
9
228
95
372
 23
23
565
25
590
 2
2
47
80
76
 17
17
405
35
508
 26
26
646
44
1057
 19
19
460
17
435
 4
4
90
95
101
 5
5
131
34
176
 18
18
452
186
528
 4
4
90
35
118
 2
2
37
12
38
 4
4
87
28
116
 1.3
1.3
31
15
37
 11
11
257
206
2667
 5
5
111
22
174
 9
9
219
101
341
 14
14
349
109
397
 4
4
92
26
88
 4
4
90
30
128
 3
3
83
23
107
 7
7
171
23
568
 6
6
151
26
191
 15
15
367
58
478
 4
4
93
38
95
 5
5
115
22
149
 3
3
81
22
86
 6
6
156
28
118
 19
19
476
24
436
 100
100
2445
36
4495
 2
2
61
72
277
 28
28
689
57
669
 5
5
127
35
85
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 32.1
32.1
47.8
100 -
-
-
100 -
-
-
98.5 -
-
-
99 98.8
98.8
99.1
98 -
-
-
- 97.9
97.9
99.1
98 38.7
38.7
53.7
97 -
-
-
- 75.3
75.3
85.5
- 34.4
34.4
51.5
96 43.5
43.5
56
- 80.6
80.6
85.9
87 -
-
-
- 58.7
58.7
74.8
61 -
-
-
- 38.3
38.3
56.2
52.5 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 32
32
53.2
- 24.3
24.3
36.1
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK14 - Q16539 |
| 2 | TAOK2 - Q9UL54 |
| 3 | RELA - Q04206 |
| 4 | MAPK12 - P53778 |
| 5 | GADD45G - O95257 |
| 6 | GADD45B - O75293 |
| 7 | SMAD7 - O15105 |
| 8 | MAP2K3 - P46734 |
| 9 | PLCB2 - Q00722 |
| 10 | GPD1 - P21695 |
| 11 | MAPK3 - P27361 |
| 12 | MAPK1 - P28482 |
Regulation
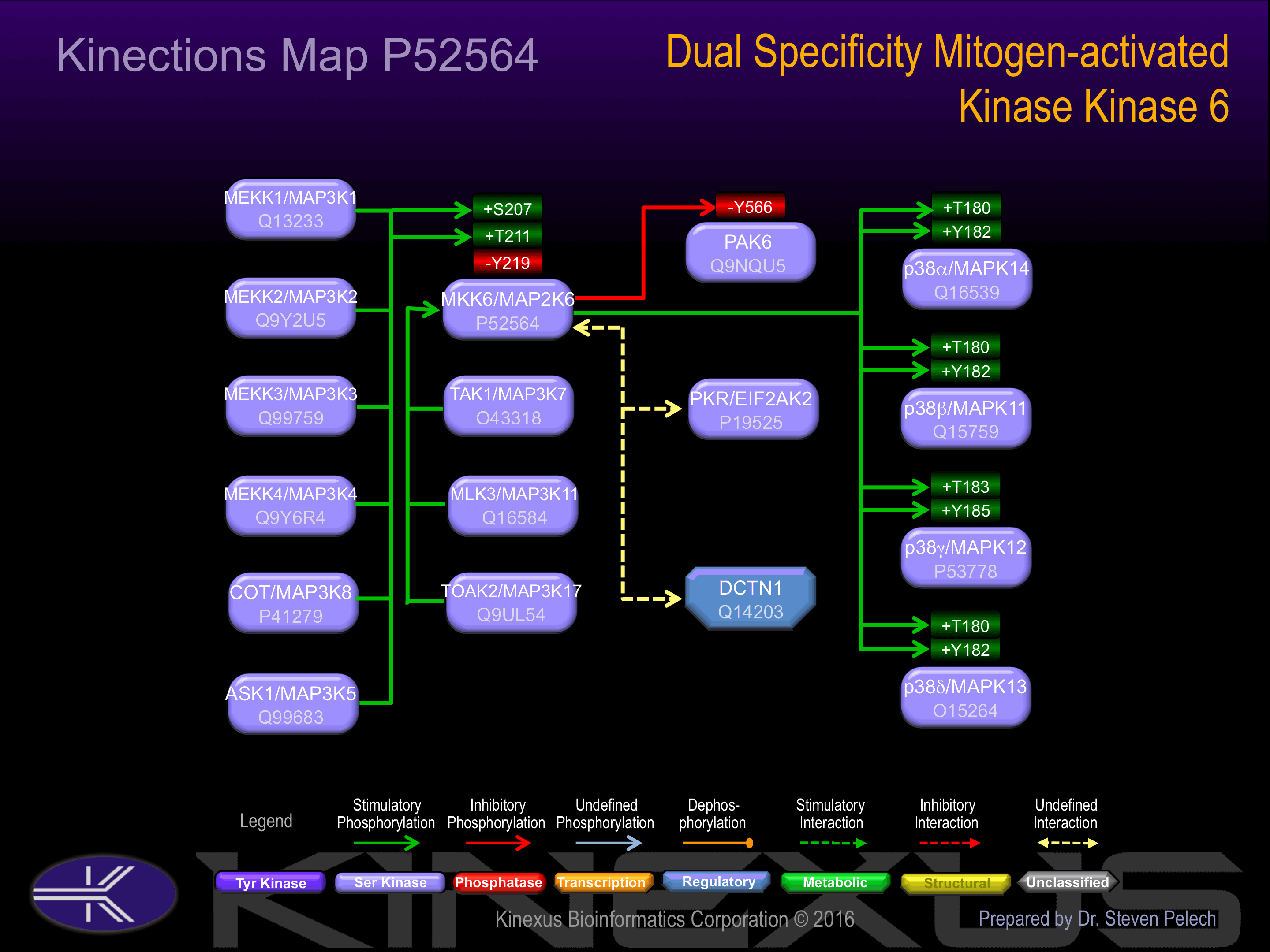
Activation:
Probably activated by dual phosphorylation on Ser-207 and Thr-211.
Inhibition:
NA
Synthesis:
Strongly activated by UV, anisomycin, and osmotic shock but not by phorbol esters, NGF or EGF.
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| COT | P41279 | S207 | ISGYLVDSVAKTIDA | + |
| ASK1 | Q99683 | S207 | ISGYLVDSVAKTIDA | + |
| MEKK1 | Q13233 | S207 | ISGYLVDSVAKTIDA | + |
| MEKK3 | Q99759 | S207 | ISGYLVDSVAKTIDA | + |
| MEKK2 | Q9Y2U5 | S207 | ISGYLVDSVAKTIDA | + |
| MEKK4 | Q9Y6R4 | S207 | ISGYLVDSVAKTIDA | + |
| ASK1 | Q99683 | T211 | LVDSVAKTIDAGCKP | + |
| MEKK1 | Q13233 | T211 | LVDSVAKTIDAGCKP | + |
| MEKK3 | Q99759 | T211 | LVDSVAKTIDAGCKP | + |
| MEKK2 | Q9Y2U5 | T211 | LVDSVAKTIDAGCKP | + |
| MEKK4 | Q9Y6R4 | T211 | LVDSVAKTIDAGCKP | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| p38a MAPK (MAPK14) | Q16539 | T180 | RHTDDEMTGYVATRW | + |
| p38a MAPK (MAPK14) | Q16539 | Y182 | TDDEMTGYVATRWYR | + |
| p38b MAPK (MAPK11) | Q15759 | T180 | RQADEEMTGYVATRW | + |
| p38b MAPK (MAPK11) | Q15759 | Y182 | ADEEMTGYVATRWYR | + |
| p38d MAPK (MAPK13) | O15264 | T180 | RHADAEMTGYVVTRW | + |
| p38d MAPK (MAPK13) | O15264 | Y182 | ADAEMTGYVVTRWYR | + |
| p38g MAPK (MAPK12) | P53778 | T183 | RQADSEMTGYVVTRW | + |
| p38g MAPK (MAPK12) | P53778 | Y185 | ADSEMTGYVVTRWYR | + |
| PAK6 | Q9NQU5 | Y566 | KSLVGTPYWMAPEVI | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
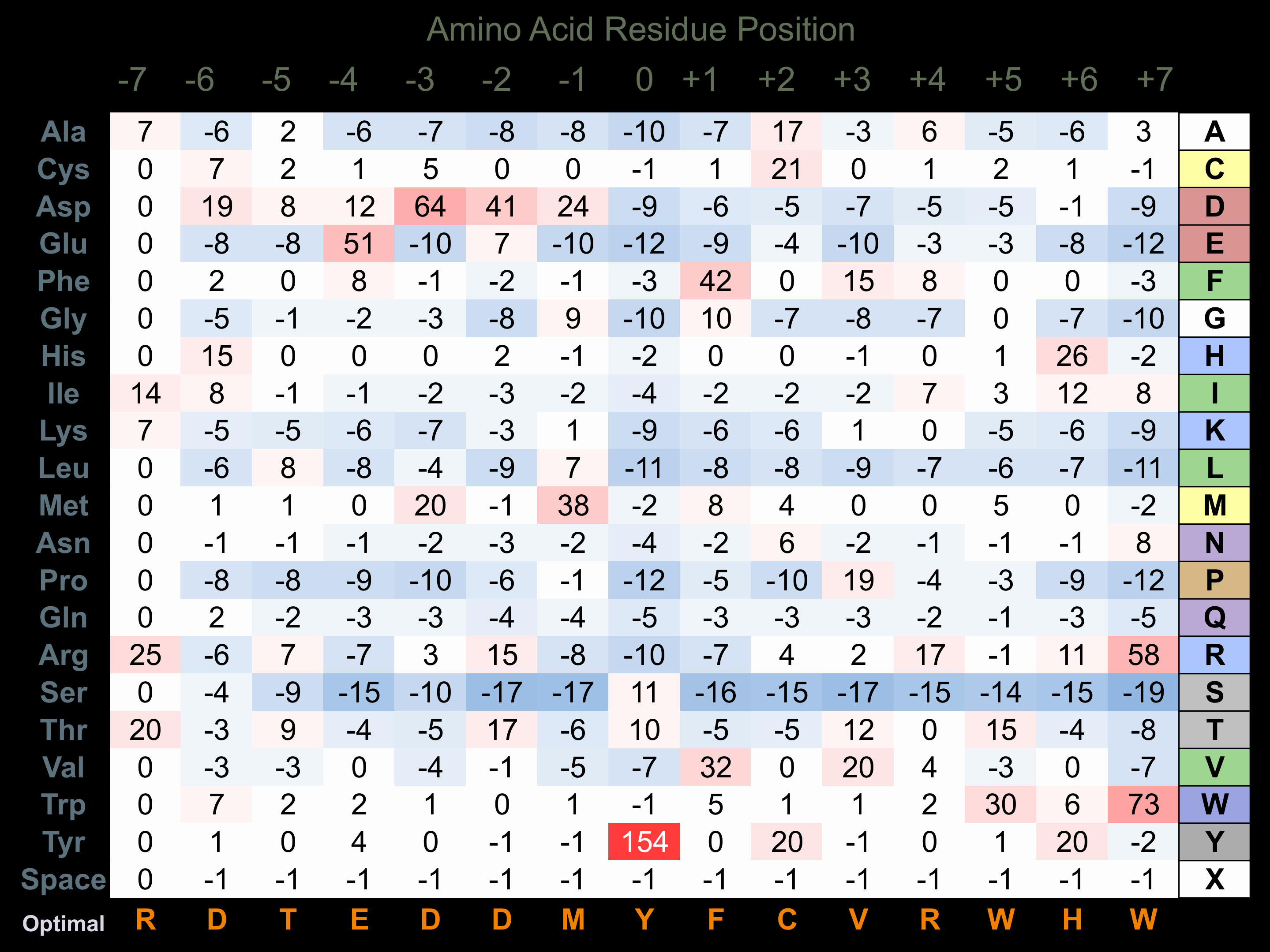
Matrix Type:
Experimentally derived from alignment of 11 known protein substrate phosphosites and 22 peptides phosphorylated by recombinant MEK6 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neonatal disorders
Specific Diseases (Non-cancerous):
Pierre Robin sequence
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer stage 2B (%CFC= -71); and Ovary adenocarcinomas (%CFC= +90, p<0.003). The COSMIC website notes an up-regulated expression score for MEK6 in diverse human cancers of 421, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 1 for this protein kinase in human cancers was 98% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. S207A and T211A mutations are associated with inactivation of the kinase. S207E and T211E are linked with constitutive activation of MEK6.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25371 diverse cancer specimens. This rate is only 12 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.2 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: K232Q (8).
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.