Nomenclature
Short Name:
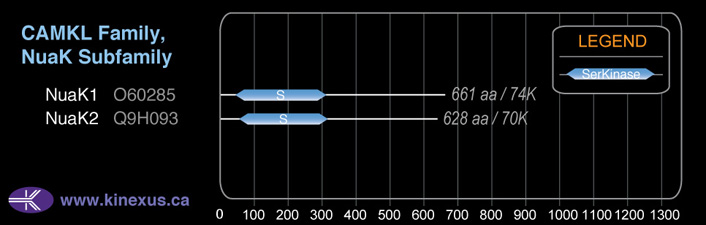
NuaK1
Full Name:
AMPK-related protein kinase 5
Alias:
- AMP-activated protein kinase family member 5
- ARK5
- NUAK family, SNF1-like kinase, 1
- NUAK2
- Probable KIAA0537
- EC 2.7.11.1
- KIAA053
- KIAA0537
- Kinase NuaK1
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
NuaK
Structure
Mol. Mass (Da):
74,305
# Amino Acids:
661
# mRNA Isoforms:
2
mRNA Isoforms:
74,305 Da (661 AA; O60285); 23,824 Da (208 AA; O60285-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 55 | 306 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
R284.
Serine phosphorylated:
S22, S145, S388, S444, S445, S455, S459, S476, S480, S499, S516, S518, S562, S564, S600+.
Threonine phosphorylated:
T211+, T356.
Tyrosine phosphorylated:
Y141, Y143, Y565.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 12
12
769
16
782
 2
2
148
10
39
 1
1
67
7
87
 17
17
1153
60
787
 12
12
765
14
666
 16
16
1068
52
2237
 5
5
347
19
571
 9
9
578
33
946
 5
5
302
10
229
 2
2
129
51
67
 2
2
107
20
77
 11
11
744
101
758
 1.1
1.1
74
18
63
 3
3
184
9
98
 1.2
1.2
77
16
59
 1.3
1.3
84
8
37
 2
2
138
105
59
 1
1
67
14
57
 4
4
239
49
126
 15
15
990
56
784
 4
4
232
16
144
 0.6
0.6
43
17
43
 1
1
66
8
67
 1
1
63
14
54
 1.4
1.4
95
16
73
 13
13
835
42
1343
 0.9
0.9
57
21
59
 1.3
1.3
87
14
61
 3
3
211
14
154
 2
2
152
14
45
 7
7
432
18
313
 100
100
6616
27
8345
 5
5
309
43
740
 12
12
768
26
668
 1
1
69
22
97
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.8
100 97.9
97.9
98
99 -
-
-
95 -
-
-
- 91.7
91.7
94.4
92 -
-
-
- 90.9
90.9
93.8
91.5 55.7
55.7
68.1
91 -
-
-
- 84.3
84.3
88
- 26.2
26.2
42.9
83 48.6
48.6
61.4
- 23.6
23.6
36.2
69 -
-
-
- 24.4
24.4
33.6
- -
-
-
- 22.5
22.5
35.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 31.2
31.2
49.3
- 26.8
26.8
46.1
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CASP6 - P55212 |
| 2 | SIK1 - P57059 |
| 3 | MARK3 - P27448 |
| 4 | BRSK2 - Q8IWQ3 |
| 5 | MARK1 - Q9P0L2 |
| 6 | SIK2 - Q9H0K1 |
| 7 | MARK4 - Q96L34 |
| 8 | NUAK2 - Q9H093 |
| 9 | BRSK1 - Q8TDC3 |
| 10 | MARK2 - Q7KZI7 |
| 11 | PRKAA1 - Q13131 |
| 12 | SIK3 - Q9Y2K2 |
| 13 | PRKAG1 - P54619 |
| 14 | HSP90AA1 - P07900 |
| 15 | HSPA1A - P08107 |
Regulation
Activation:
Activated by phosphorylation at Thr-211 and Ser-600.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
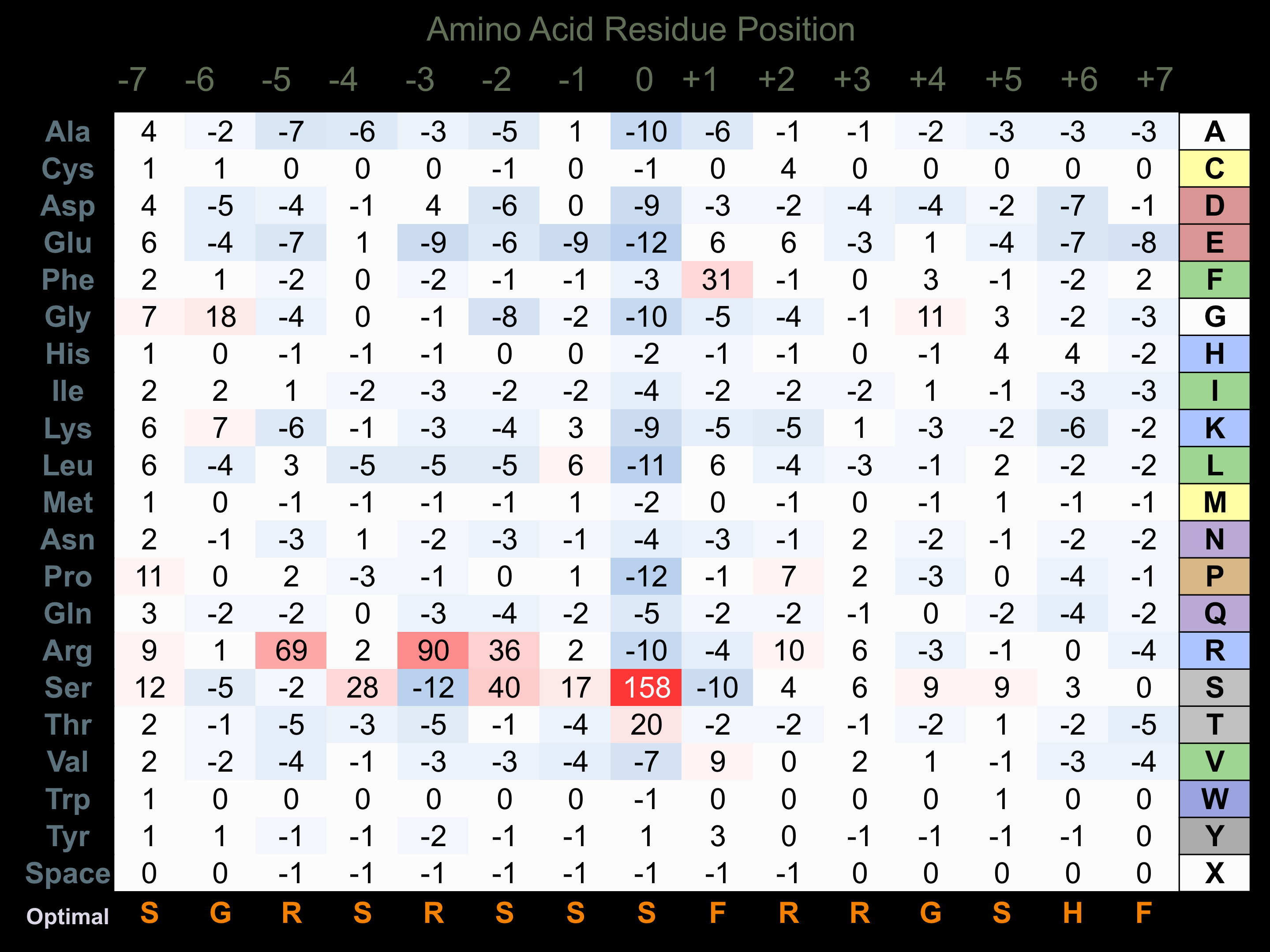
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Developmental disorders
Specific Diseases (Non-cancerous):
Omphalocele
Comments:
Omphalocele is a rare syndrome typically characterized by the liver, intestines, and sometimes other organs being located outside of the abdomen. These organs are surrounded in a sack, but can still lead to high rates of mortality. The tissues affected by Omphalocele can include the liver, skin, and heart. A K84A mutation in NuaK1 can lead to inhibited kinase phosphotransferase activity and can lead to senescence of the cell (consisting of biologically old cell characteristics).
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +84, p<0.034); Brain glioblastomas (%CFC= +863, p<0.004); Brain oligodendrogliomas (%CFC= +825, p<0.084); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -72, p<0.016); Colorectal adenocarcinomas (early onset) (%CFC= +116, p<0.002); Oral squamous cell carcinomas (OSCC) (%CFC= +742, p<0.001); Ovary adenocarcinomas (%CFC= +190, p<0.002); Pituitary adenomas (ACTH-secreting) (%CFC= +68); Skin fibrosarcomas (%CFC= -45); Skin melanomas - malignant (%CFC= -82, p<0.002); and Vulvar intraepithelial neoplasia (%CFC= -67, p<0.0002).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A T211A NuaK1 mutation can inhibit LKB1-mediated activation and phosphorylation of NuaK1, while ablating induction of senescence. The I400K and L401K mutations can lead to interrupted NuaK1 complex with PPP1CB, and inhibiting regulation of PP1 activity. Phosphorylation via AKT1 can be prevented through an S600A mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 24434 diverse cancer specimens. This rate is only 37 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 3 in 20,034 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

