Nomenclature
Short Name:
PKR
Full Name:
Interferon-induced, double-stranded RNA-activated protein kinase
Alias:
- ADRB2
- EIF2AK2
- Eukaryotic translation initiation factor 2-alpha kinase 2
- Interferon-inducible RNA-dependent protein kinase
- P1,eIF-2A protein kinase
- TIK
- E2AK2
- EC 2.7.11.1
- EIF2aK
- EIF2AK1
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
PEK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
62,094
# Amino Acids:
551
# mRNA Isoforms:
2
mRNA Isoforms:
62,094 Da (551 AA; P19525); 57,391 Da (510 AA; P19525-2)
4D Structure:
Homodimer. Interacts with STRBP By similarity. Interacts with DNAJC3. Inhibited by direct interaction with viral proteins such as HCV E2, HCV NS5A and influenza A NS1. Activated by the interaction with HIV-1 Tat.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
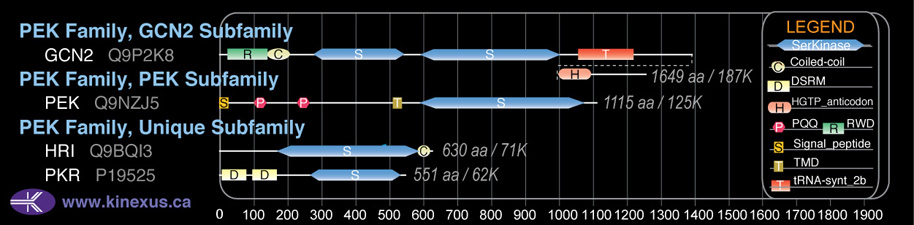
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2.
Methylated:
K61, K69.
Serine phosphorylated:
S6, S33, S83+, S92, S179, S181, S242+, S456-, S542.
Threonine phosphorylated:
T88+, T89+, T90+, T255+, T258+, T446+, T451-, T535.
Tyrosine phosphorylated:
Y101+, Y142, Y162+, Y257, Y293+, Y454.
Ubiquitinated:
K268, K299, K304, K385, K388, K400, K408, K416, K426, K429, K440, K509, K517, K521.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 34
34
1126
35
955
 5
5
171
18
125
 11
11
363
18
341
 22
22
716
140
1142
 26
26
858
34
668
 7
7
230
99
331
 11
11
367
45
586
 100
100
3280
66
5641
 19
19
637
17
495
 13
13
427
131
1041
 10
10
321
45
442
 23
23
754
212
680
 8
8
260
40
282
 7
7
215
15
136
 7
7
227
23
369
 6
6
193
21
131
 14
14
467
297
1359
 9
9
311
29
337
 4
4
128
123
109
 20
20
657
137
629
 10
10
317
37
354
 11
11
372
41
513
 11
11
377
28
728
 4
4
123
29
135
 11
11
345
37
416
 73
73
2406
93
3808
 6
6
192
43
192
 16
16
522
29
1548
 10
10
332
29
388
 3
3
94
42
65
 30
30
984
24
795
 17
17
565
41
702
 3
3
102
86
162
 31
31
1008
83
852
 5
5
158
48
182
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.5
98.5
99.3
99 80.5
80.5
87.7
81 -
-
-
62 -
-
-
61 21.1
21.1
33.3
65 -
-
-
- 58.3
58.3
71.3
62 57.5
57.5
70.8
63 -
-
-
- -
-
-
- 38.1
38.1
56.1
43 34.8
34.8
55.2
40 29.3
29.3
45.2
41 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | ILF3 - Q12906 |
| 2 | SMAD2 - Q15796 |
| 3 | TARBP2 - Q15633 |
| 4 | NPM1 - P06748 |
| 5 | PRKRA - O75569 |
| 6 | EIF2S1 - P05198 |
| 7 | TP53 - P04637 |
| 8 | PPP1CA - P62136 |
| 9 | PTGES3 - Q15185 |
| 10 | PRKRIR - O43422 |
| 11 | STAT3 - P40763 |
| 12 | METAP2 - P50579 |
| 13 | PPP2R5A - Q15172 |
| 14 | PDGFRB - P09619 |
| 15 | CDC42 - P60953 |
Regulation
Activation:
Besides double stranded RNA (dsRNA), heparin is a potent activator of the kinase. Binding to dsRNA is required for dimerization leading to autophosphorylation in the activation loop and stimulation of function. Activity is markedly stimulated by manganese ions. Autophosphorylation at Ser-83, Thr-88, Thr-89, Thr-90, Tyr-101, Tyr-162, Ser-242, Thr-255, Thr-258, Tyr-293 Thr-446, Thr-451, and Ser-456 increase the phosphotransferase activity of EIF2AK2.
Inhibition:
Inhibited by vaccinia virus protein E3, probably via dsRNA sequestering.
Synthesis:
By interferon.
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKR | P19525 | S83 | NKEKKAVSPLLLTTT | + |
| PKR | P19525 | T88 | AVSPLLLTTTNSSEG | + |
| PKR | P19525 | T89 | VSPLLLTTTNSSEGL | + |
| PKR | P19525 | T90 | SPLLLTTTNSSEGLS | + |
| PKR | P19525 | Y101 | EGLSMGNYIGLINRI | + |
| PKR | P19525 | Y162 | QLAAKLAYLQILSEE | + |
| PKR | P19525 | S242 | NQRKAKRSLAPRFDL | + |
| PKR | P19525 | T255 | DLPDMKETKYTVDKR | + |
| PKR | P19525 | T258 | DMKETKYTVDKRFGM | + |
| PKR | P19525 | Y293 | HRIDGKTYVIKRVKY | + |
| JAK1 | P23458 | Y293 | HRIDGKIYVIKRVKY | + |
| PKR | P19525 | T446 | LKNDGKRTRSKGTLR | + |
| ERK2 | P28482 | T451 | KRTRSKGTLRYMSPE | - |
| RSK2 | P51812 | T451 | KRTRSKGTLRYMSPE | - |
| p38a | Q16539 | T451 | KRTRSKGTLRYMSPE | - |
| PKR | P19525 | T451 | KRTRSKGTLRYMSPE | - |
| PKR | P19525 | S456 | KGTLRYMSPEQISSQ | - |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
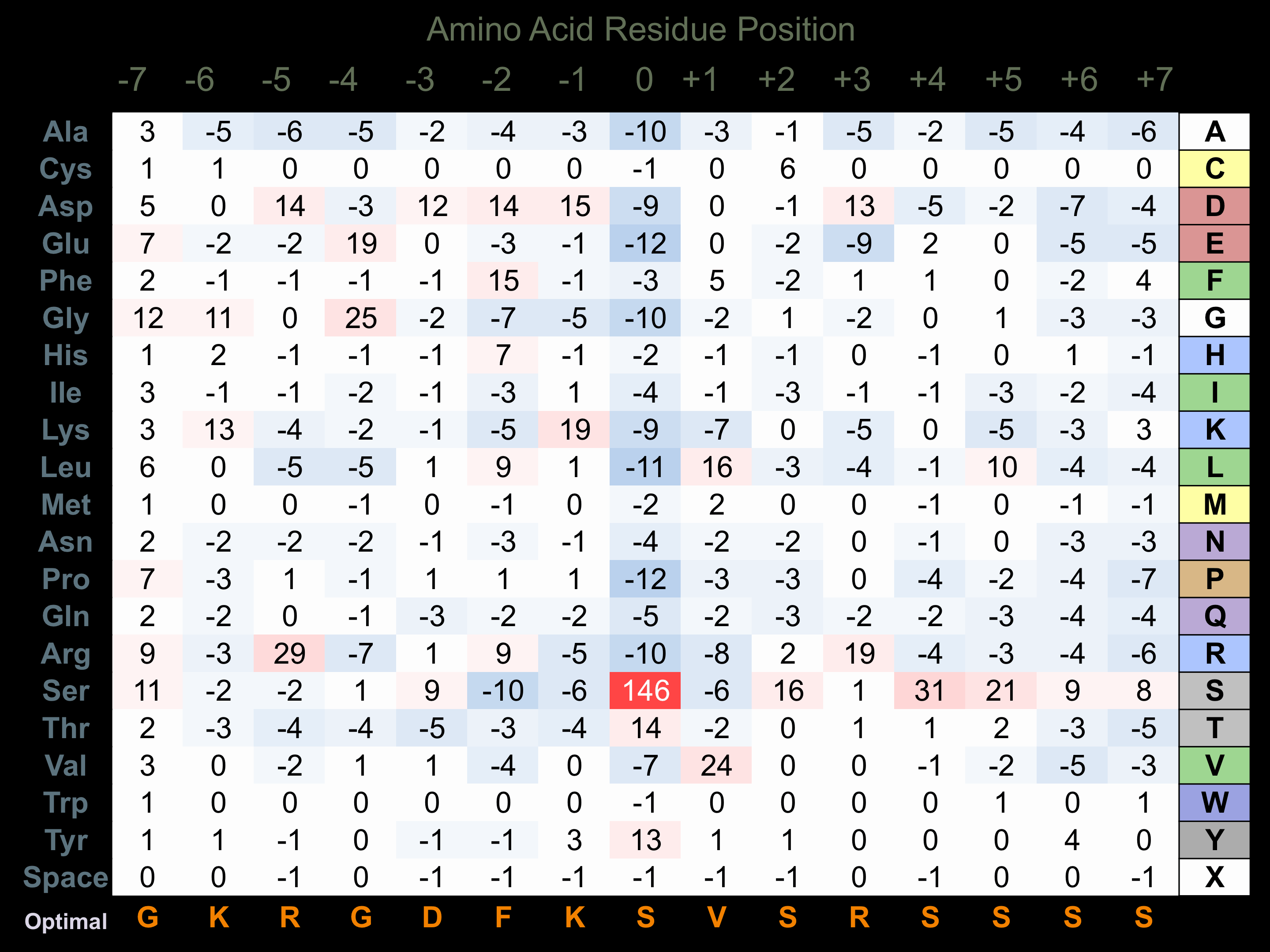
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Infectious disease; and nephrological disorder
Specific Diseases (Non-cancerous):
Herpes simplex; Fanconi Anemia, Complementation Group C; Hepatitis D; Birt-Hogg-Dube syndrome; Rift Valley Fever; Parainfluenza virus Type 3
Comments:
PKR is a protein-serine/threonine kinase that has an antiviral activity on various ranges of DNA and RNA viruses such as hepatitis C virus, hepatitis B virus, measles virus, and herpes simplex virus 1. It has a role in p53-mediated responses to genotoxic stress. Since the protein has a crucial role for inflammation activation, it can be a potential therapeutic target to treat inflammation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +65, p<0.025); Bladder carcinomas (%CFC= +69, p<0.0006); Brain oligodendrogliomas (%CFC= -63, p<0.022); Cervical cancer (%CFC= -60, p<0.0001); Cervical cancer stage 2B (%CFC= +96, p<0.035); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -57, p<0.0004); Oral squamous cell carcinomas (OSCC) (%CFC= +169, p<0.0001); and Prostate cancer - primary (%CFC= -93, p<0.0001). The COSMIC website notes an up-regulated expression score for PKR in diverse human cancers of 490, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 7 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25183 diverse cancer specimens. This rate is only -9 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.45 % in 602 endometrium cancers tested; 0.23 % in 805 skin cancers tested; 0.17 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,466 cancer specimens
Comments:
Only 4 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

