Nomenclature
Short Name:
JAK1
Full Name:
Tyrosine-protein kinase JAK1
Alias:
- EC 2.7.10.2
- JAK1A
- Janus kinase 1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
JakA
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
133277
# Amino Acids:
1154
# mRNA Isoforms:
1
mRNA Isoforms:
133,277 Da (1154 AA; P23458)
4D Structure:
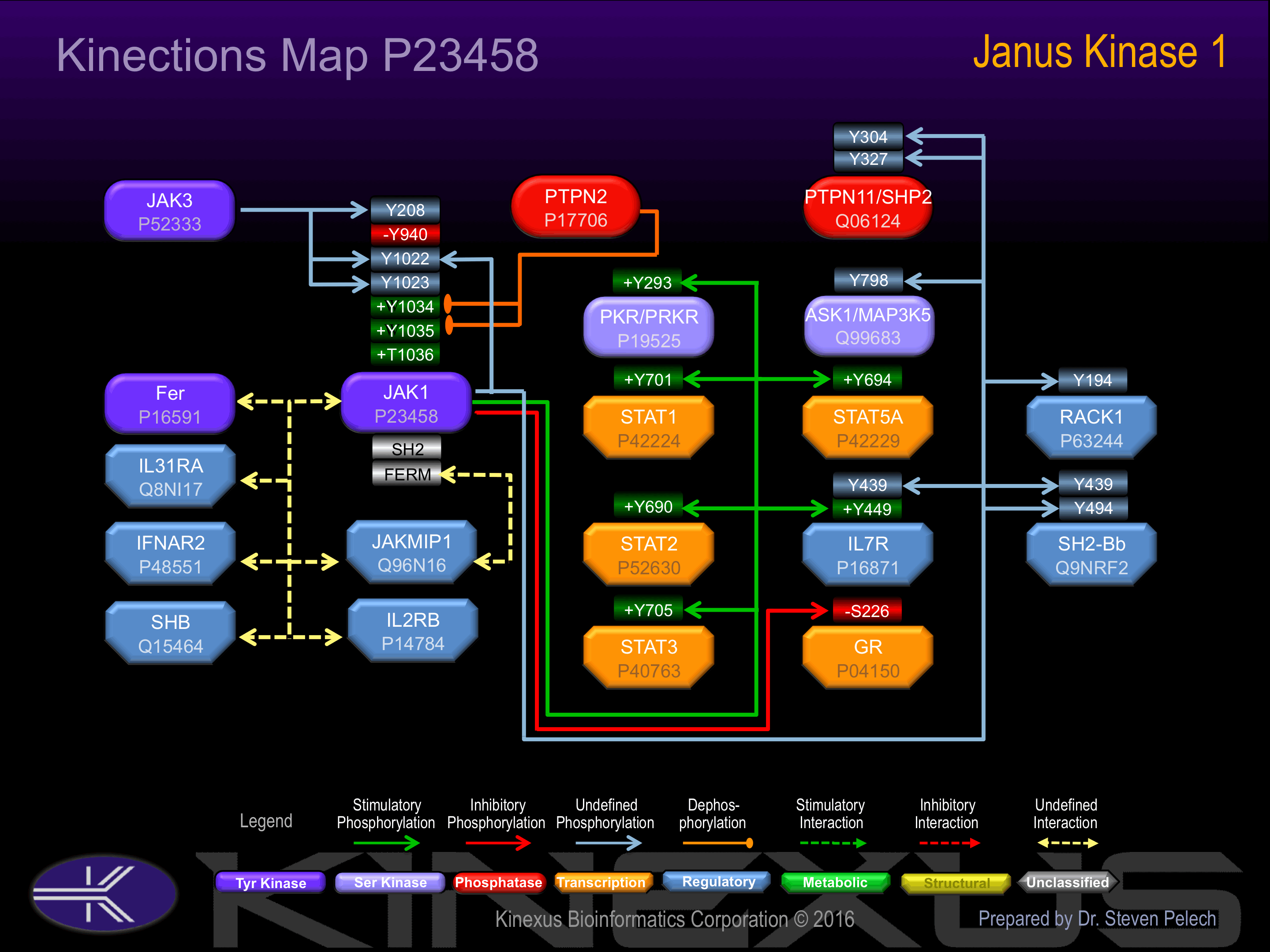
Interacts with IL31RA, JAKMIP1 and SHB.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
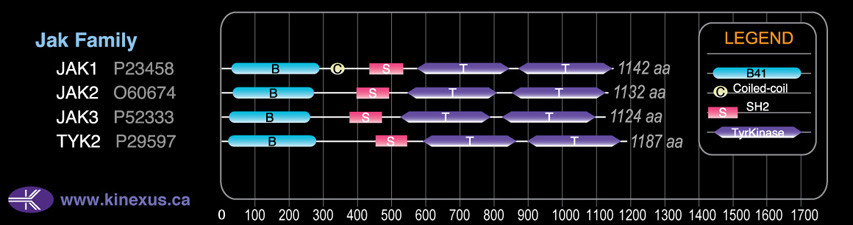
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
M1, K269, K1109, K1112.
Serine phosphorylated:
S216, S228, S333, S574, S738, S857, S909, S1043\.
Threonine phosphorylated:
T687, T688, T1030, T1036+, T1107.
Tyrosine phosphorylated:
Y3, Y208, Y217, Y220, Y412, Y568-, Y605, Y940-, Y967, Y993, Y1022, Y1023, Y1034+, Y1035+, Y1125.
Ubiquitinated:
K213, K227, K245, K249, K269, K736, K1109.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1647
46
810
 11
11
180
19
158
 44
44
729
24
652
 20
20
330
202
561
 57
57
942
55
683
 17
17
274
92
306
 25
25
412
77
597
 49
49
800
64
1277
 58
58
948
10
802
 19
19
309
146
375
 18
18
296
47
325
 53
53
867
206
763
 59
59
978
35
890
 23
23
379
15
325
 24
24
392
44
441
 10
10
171
35
222
 19
19
308
319
1802
 25
25
407
35
432
 23
23
372
121
488
 58
58
963
181
695
 30
30
487
43
517
 49
49
815
45
929
 28
28
466
33
522
 36
36
591
35
801
 48
48
785
43
872
 57
57
934
145
1214
 46
46
762
38
706
 23
23
372
35
389
 38
38
621
35
696
 5
5
75
42
66
 94
94
1544
18
195
 78
78
1281
41
4581
 39
39
636
117
916
 58
58
948
161
691
 74
74
1220
87
2701
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 20.5
20.5
35.4
100 99.5
99.5
99.7
99.5 -
-
-
97 -
-
-
97 97.5
97.5
98.6
98 -
-
-
- 94.6
94.6
96.8
95 43.8
43.8
62.7
95 -
-
-
- -
-
-
- 43.8
43.8
62.7
82 75.8
75.8
86.2
78 62.7
62.7
76.5
64 -
-
-
- 21.2
21.2
40
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
Phosphorylation of Tyr-940 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ASK1 (MAP3K5) | Q99683 | Y798 | IPERDSRYSQPLHEE | |
| GR | P04150 | S226 | IDENCLLSPLAGEDD | - |
| IL7R | P16871 | Y439 | VLNQEEAYVTMSSFY | |
| IL7R | P16871 | Y449 | GSNQEEAYVTMSSFY | + |
| JAK1 | P23458 | Y1022 | AIETDKEYYTVKDDR | |
| PKR (PRKR; EIF2AK2) | P19525 | Y293 | HRIDGKIYVIKRVKY | + |
| PTPN11 (SHP2) | Q06124 | Y304 | PNEPVSDYINANIIM | |
| PTPN11 (SHP2) | Q06124 | Y327 | NSKPKKSYIATQGCL | |
| RACK1 | P63244 | Y194 | NHIGHTGYLNTVTVS | |
| SH2-Bb | Q9NRF2 | Y439 | DRLSQGAYGGLSDRP | |
| SH2-Bb | Q9NRF2 | Y494 | VHPLSAPYPPLDTPE | |
| STAT1 | P42224 | Y701 | DGPKGTGYIKTELIS | + |
| STAT2 | P52630 | Y690 | NLQERRKYLKHRLIV | + |
| STAT3 | P40763 | Y705 | DPGSAAPYLKTKFIC | + |
| STAT5A | P42229 | Y694 | LAKAVDGYVKPQIKQ | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
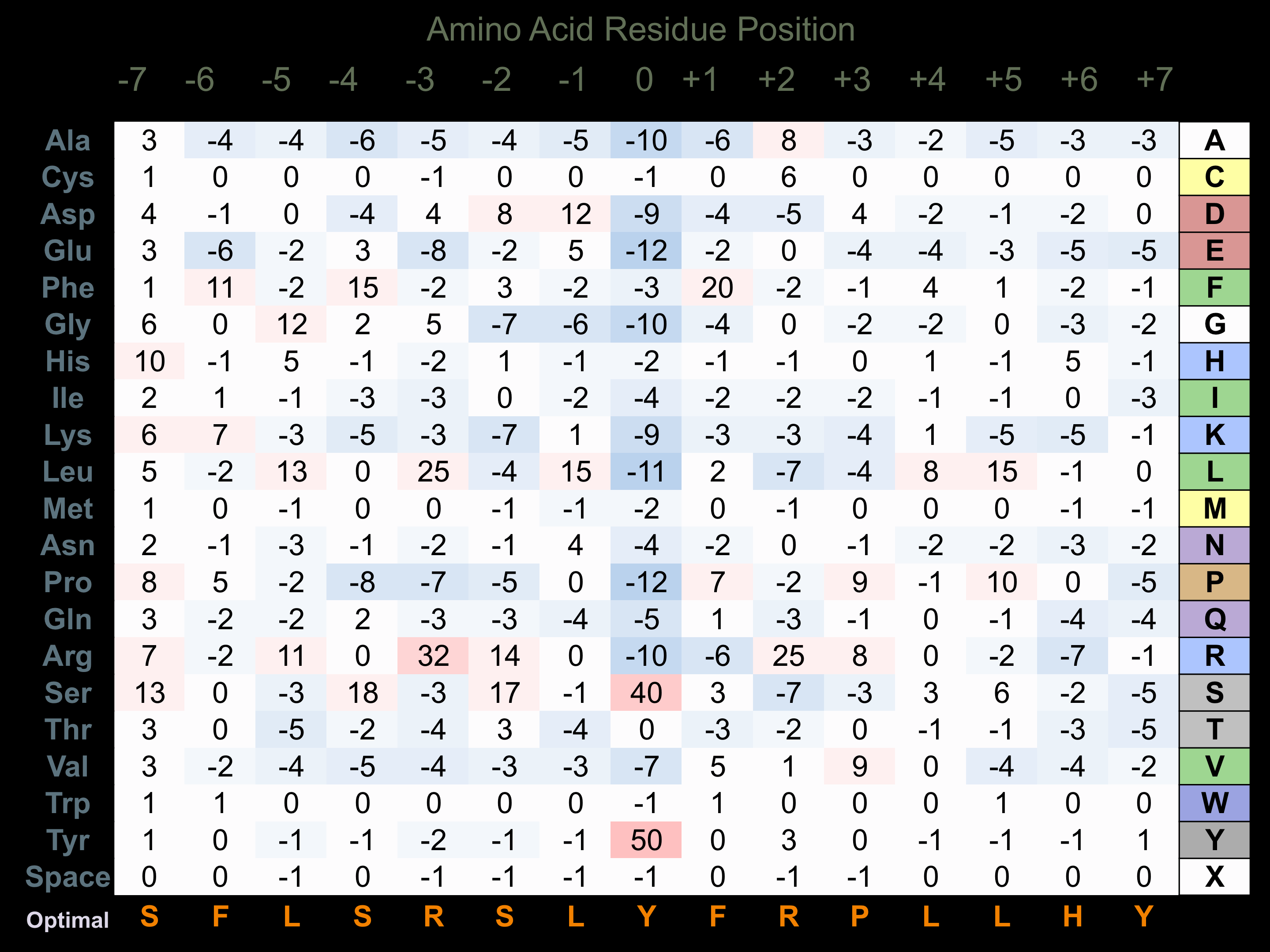
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
2
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, immune disorders
Specific Diseases (Non-cancerous):
Behcet's disease
Comments:
Behcet's disease, also known as silk road disease, is a rare immune disease characterized by small blood vessel vasculitis, mucous membrane ulceration, and ocular defects. Affected systems include the gastrointestinal tract, pulmonary, musculoskeletal, cardiovascular, and nervous system. This disease is often fatal due to aneurysms or severe neurological defects. JAK1 has been proposed as a suceptibility gene for Behcet's disease. Three single nucleotide polymorphisms (SNPs) in the JAK1 gene were associated with a signficantly increased risk for the development of Behcet's disease in Han Chinese populations, including rs2780815, rs310241, and rs3790532. When found in an individual, these SNPs were estimated to convey a risk of developing Behcet's disease of 35%, 28%, and 27%, respectively.
Specific Cancer Types:
Colorectal cancer (CRC); Acute myeloid leukemias (AML); Non-small cell lung cancer (NSCLC); Gynecologic cancer
Comments:
JAK1 may be an oncoprotein (OP). Abnormal activity of STAT3 is implicated in the oncogenesis of several cancer types, including colorectal cancer and non-small cell lung cancer. JAK1 has been implicated in promoting the survival, invasion, and migration of colorectal cancer and non-small cell lung cancer cells due to over activation of STAT transcription factors resulting in the aberrant expression of certain downstream genes, such as Bcl-2, p21, p27, E-cadherin, VEGF, and MMPs. In addition, somatic mutations in the JAK1 gene have been observed in patients with acute myeloid leukemia (AML), including both T478S and V623A substitution mutations. The mutant JAK1 proteins displayed significantly enhanced activation of downstream STAT1 in response to type 1 interferon treatment, as well as increased activation of multiple dowstream intracellular signalling pathways. Truncating mutations of JAK1 have been observed in several cancer types, with 68% of the specimens coming from gynecologic cancers. Within the JAK1 gene sequence, mutations are clustered at the K142, P430, and K860 residues and induce frameshift mutations. Cancer cells with mutant truncated JAK1 protein are defective in IFN-gamma induced expression of LMP2 and TAP1, which inhibits the presentation of tumour antigens to be recognized by the immune system. Therefore, these truncating mutations are hypothesized to promote the ability of tumour cells to evade the immune system, enabling tumour survival.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +46, p<0.026); Bladder carcinomas (%CFC= +109, p<0.0001); Cervical cancer stage 2A (%CFC= +147, p<0.008); Cervical cancer stage 2B (%CFC= +124, p<0.103); Classical Hodgkin lymphomas (%CFC= -48, p<0.083); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -91, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +84, p<0.0006); Ovary adenocarcinomas (%CFC= -57, p<0.017); Pituitary adenomas (ACTH-secreting) (%CFC= +149); and Skin melanomas - malignant (%CFC= -47, p<0.0001). The COSMIC website notes an up-regulated expression score for JAK1 in diverse human cancers of 256, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 156 for this protein kinase in human cancers was 2.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 30277 diverse cancer specimens. This rate is only 20 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.36 % in 1393 large intestine cancers tested; 0.32 % in 602 endometrium cancers tested; 0.22 % in 683 stomach cancers tested; 0.14 % in 805 skin cancers tested; 0.11 % in 5026 haematopoietic and lymphoid cancers tested; 0.1 % in 2069 lung cancers tested; 0.08 % in 1654 liver cancers tested; 0.07 % in 1709 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R724H (10); S703I (7); V658F (7).
Comments:
Broad distribution of mutation sites with many point mutations, deletions, insertions and at least three complex mutations across the entire protein.