Nomenclature
Short Name:
FGFR4
Full Name:
Fibroblast growth factor receptor 4
Alias:
- CD334
- EC 2.7.10.1
- JTK2
- MGC20292
- MPK11
- TKF
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
FGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
87,954
# Amino Acids:
802
# mRNA Isoforms:
3
mRNA Isoforms:
87,954 Da (802 AA; P22455); 83,452 Da (762 AA; P22455-2); 64,640 Da (592 AA; P22455-3)
4D Structure:
Interacts with KLB
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
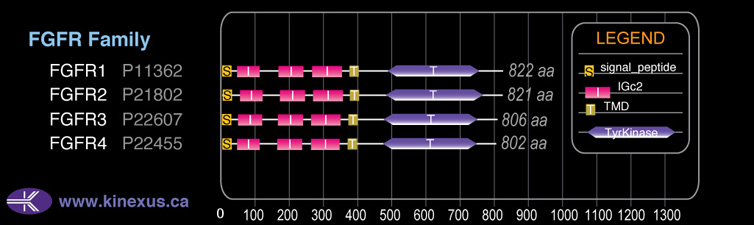
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N112, N258, N290, N311, N322.
Serine phosphorylated:
S410, S419, S439, S440, S441, S519, S573.
Threonine phosphorylated:
T167, T368.
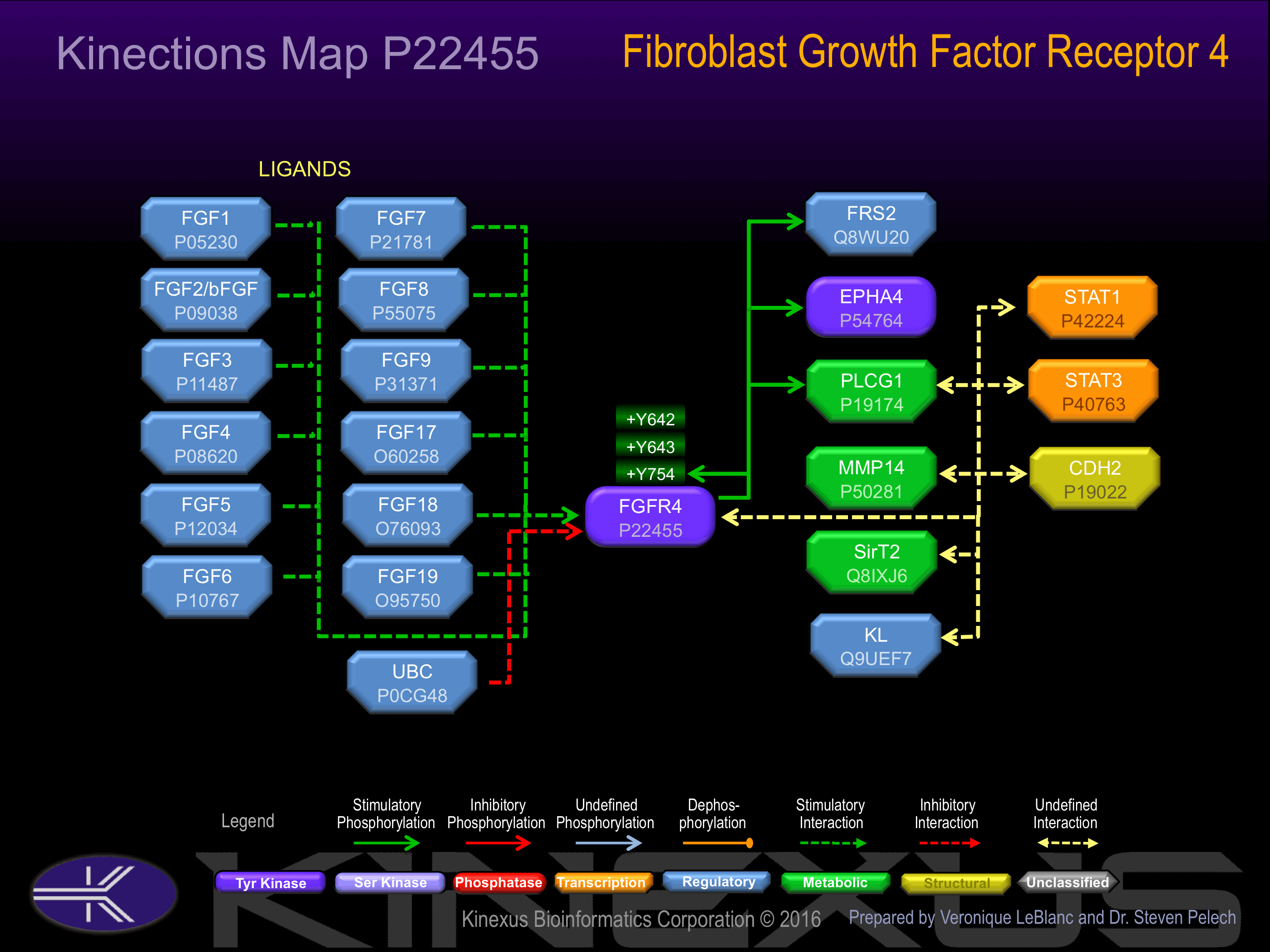
Tyrosine phosphorylated:
Y642+, Y643+, Y754.
Ubiquitinated:
K627.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 64
64
756
41
1027
 3
3
31
17
23
 5
5
57
33
76
 42
42
492
163
2236
 47
47
560
36
340
 27
27
319
100
1131
 11
11
130
51
374
 100
100
1183
67
2798
 33
33
390
17
328
 9
9
102
106
104
 16
16
188
52
164
 57
57
677
208
609
 5
5
54
55
100
 6
6
73
12
74
 12
12
146
45
139
 3
3
30
25
24
 4
4
53
150
56
 6
6
70
36
106
 3
3
30
108
57
 25
25
297
155
271
 5
5
57
46
73
 10
10
121
50
153
 9
9
101
44
112
 6
6
76
42
133
 9
9
106
38
152
 84
84
994
115
2333
 7
7
78
57
187
 8
8
92
44
140
 6
6
75
43
103
 5
5
54
28
39
 24
24
288
24
287
 43
43
507
46
596
 0.2
0.2
2
58
1
 55
55
655
109
588
 3
3
36
79
40
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 89.2
89.2
89.7
99 97.4
97.4
98.4
97 -
-
-
93 -
-
-
- 86.7
86.7
88.6
93 -
-
-
- 89.8
89.8
92.5
91 88.8
88.8
91.7
91 -
-
-
- -
-
-
- 61.5
61.5
73
73 63.9
63.9
74.6
- 56.4
56.4
68.7
66 -
-
-
- 35.5
35.5
52.1
- -
-
-
- 31
31
46.1
- 34.3
34.3
49.3
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | FGF4 - P08620 |
| 2 | FGF5 - P12034 |
| 3 | FGF2 - P09038 |
| 4 | FGF6 - P10767 |
| 5 | FGF19 - O95750 |
| 6 | FGF8 - P55075 |
| 7 | CDH2 - P19022 |
| 8 | FGF18 - O76093 |
| 9 | EPHA4 - P54764 |
| 10 | STAT3 - P40763 |
| 11 | KGFLP1 - Q2TVT4 |
| 12 | FGF7 - P21781 |
| 13 | KGFLP2 - Q2TVT3 |
Regulation
Activation:
Activated by binding of fibroblast growth factor (FGF), which induces dimerization and autophosphorylation.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
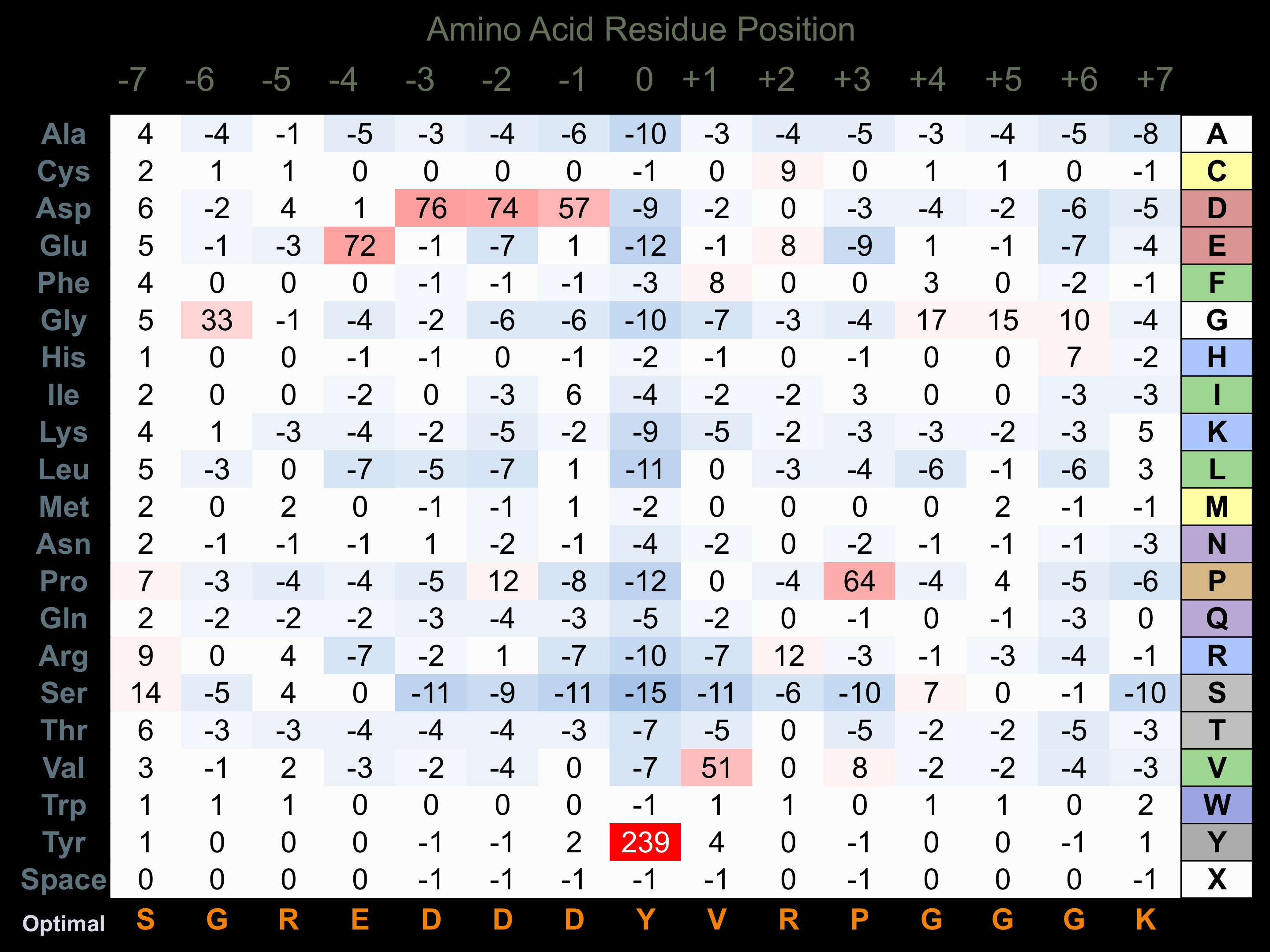
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, bone disorders
Specific Diseases (Non-cancerous):
Achondroplasia (ACH)
Comments:
Achondroplasia is a bone disease that is characterized by the failed conversion of cartilage into bone during skeletal development. Symptoms of this disease include dwarfism, limited range of motion at the joints, large-size head, small fingers, and normal intelligence.
Specific Cancer Types:
Prostate cancer; Alveolar rhabdomyosarcomas (ARMS); Breast fibroadenomas; Gliomatosis cerebri; Soft tissue sarcoma; Anaplastic astrocytoma
Comments:
FGFR4 may be an oncoprotein (OP). Defects in FGFR4 have been linked to Alveolar Rhabdomyosarcoma (ARMS), which is a rare sarcoma disease which is characterised by its alveolar-like appearance. Breast Fibroadenoma is a rare form of breast cancer. Gliomatosis Cerebri is a rare form of scattered and widespread brain tumour that is difficult to treat. Anaplastic Astrocytoma is a rare neuronal cancer but can also be associated with brain, spine cord, and bone tissues.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer stage 2A (%CFC= -46, p<0.009); Gastric cancer (%CFC= +198, p<0.0001); and Lung adenocarcinomas (%CFC= -67, p<0.0001). The COSMIC website notes an up-regulated expression score for FGFR4 in diverse human cancers of 480, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. The kinase phosphotransferase activity of FGFR4 can be inhibited with a K503R mutation. Interaction with PLCG1 can be lost with a Y754F mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25562 diverse cancer specimens. This rate is only 18 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.5 % in 805 skin cancers tested; 0.41 % in 1093 large intestine cancers tested; 0.23 % in 602 endometrium cancers tested; 0.23 % in 589 stomach cancers tested; 0.13 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: V510L (4).
Comments:
Only 5 deletions, 1 insertion and 1 complex mutation are noted on the COSMIC website.