Nomenclature
Short Name:
PKCh
Full Name:
Protein kinase C, eta type
Alias:
- EC 2.7.11.13
- NPKC-eta
- PKCL
- PKC-L
- PRKCH
- PRKCL; Protein kinase C, eta; Protein kinase C, eta type
- Kinase PKC-eta
- KPCL
- MGC5363
- MGC26269
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKC
SubFamily:
Eta
Structure
Mol. Mass (Da):
77828
# Amino Acids:
683
# mRNA Isoforms:
2
mRNA Isoforms:
77,828 Da (683 AA; P24723); 59,521 Da (522 AA; P24723-2)
4D Structure:
Interacts with DGKQ
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
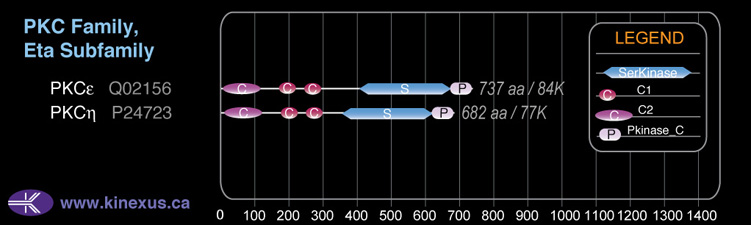
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S28, S32, S249, S317, S320, S326, S365, S599, S675+.
Threonine phosphorylated:
T25, T183, T513+, T601, T656+.
Tyrosine phosphorylated:
Y94, Y520-, Y676.
Ubiquitinated:
K36, K321.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 65
65
978
32
1027
 5
5
69
18
41
 6
6
87
15
68
 18
18
276
97
439
 53
53
803
28
709
 8
8
116
79
87
 12
12
182
34
345
 47
47
714
49
2014
 37
37
561
20
479
 8
8
117
89
126
 5
5
79
37
52
 52
52
784
181
717
 21
21
320
37
142
 4
4
60
14
39
 6
6
83
18
71
 5
5
75
15
59
 4
4
56
201
53
 13
13
195
26
110
 8
8
114
93
67
 48
48
718
112
693
 5
5
75
31
49
 22
22
335
35
215
 6
6
88
25
81
 3
3
39
27
35
 21
21
318
31
194
 62
62
930
67
1732
 15
15
221
43
109
 8
8
118
27
76
 9
9
130
27
83
 4
4
62
28
61
 100
100
1507
24
699
 37
37
558
30
472
 20
20
301
92
518
 61
61
918
57
804
 12
12
177
35
252
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
100
100 65.7
65.7
80.6
95 -
-
-
98 -
-
-
- 97.6
97.6
99.8
98 -
-
-
- 97.5
97.5
98.9
98 97.3
97.3
98.9
97 -
-
-
- -
-
-
- 86.2
86.2
93.1
86.5 75.4
75.4
85.8
- 35.5
35.5
52.7
68 -
-
-
- 41.8
41.8
57.6
- -
-
-
- 55
55
71.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 26.7
26.7
39.3
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDK2 - P24941 |
Regulation
Activation:
Phosphorylation of Thr-513 by PDK1 induces preactivation of PKC-eta, which permits autophosphorylation at T656 and T675, which are required for full activation.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AID | Q9GZX7 | T140 | GVQIAIMTFKDYFYC | |
| GSK3a | P49840 | S21 | SGRARTSSFAEPGGG | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| GSTP1 | P09211 | S184 | SAYVGRLSARPKLKA | |
| GSTP1 | P09211 | S42 | VETWQEGSLKASCLY | |
| ITGB2 | P05107 | S745 | FEKEKLKSQWNNDNP | |
| ITGB2 | P05107 | T758 | NPLFKSATTTVMNPK | |
| mGluR5 | P41594 | S840 | VRSAFTTSTVVRMHV | |
| PKCh (PRKCH) | P24723 | S675 | QDEFRNFSYVSPELQ | + |
| PKCh (PRKCH) | P24723 | T656 | IKEEPVLTPIDEGHL | + |
| PKD1 (PRKCM) | Q15139 | S738 | ARIIGEKSFRRSVVG | + |
| PKD1 (PRKCM) | Q15139 | S742 | GEKSFRRSVVGTPAY | + |
| PKD2 (PRKD2) | Q9BZL6 | S706 | ARIIGEKSFRRSVVG | + |
| PKD2 (PRKD2) | Q9BZL6 | S710 | GEKSFRRSVVGTPAY | + |
| PKD2 (PRKD2) | Q9BZL6 | S876 | QGLAERISVL_____ | + |
| PTPN11 (SHP2) | Q06124 | S580 | CAEMREDSARVYENV | |
| PTPN11 (SHP2) | Q06124 | S595 | GLMQQQKSFR_____ |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
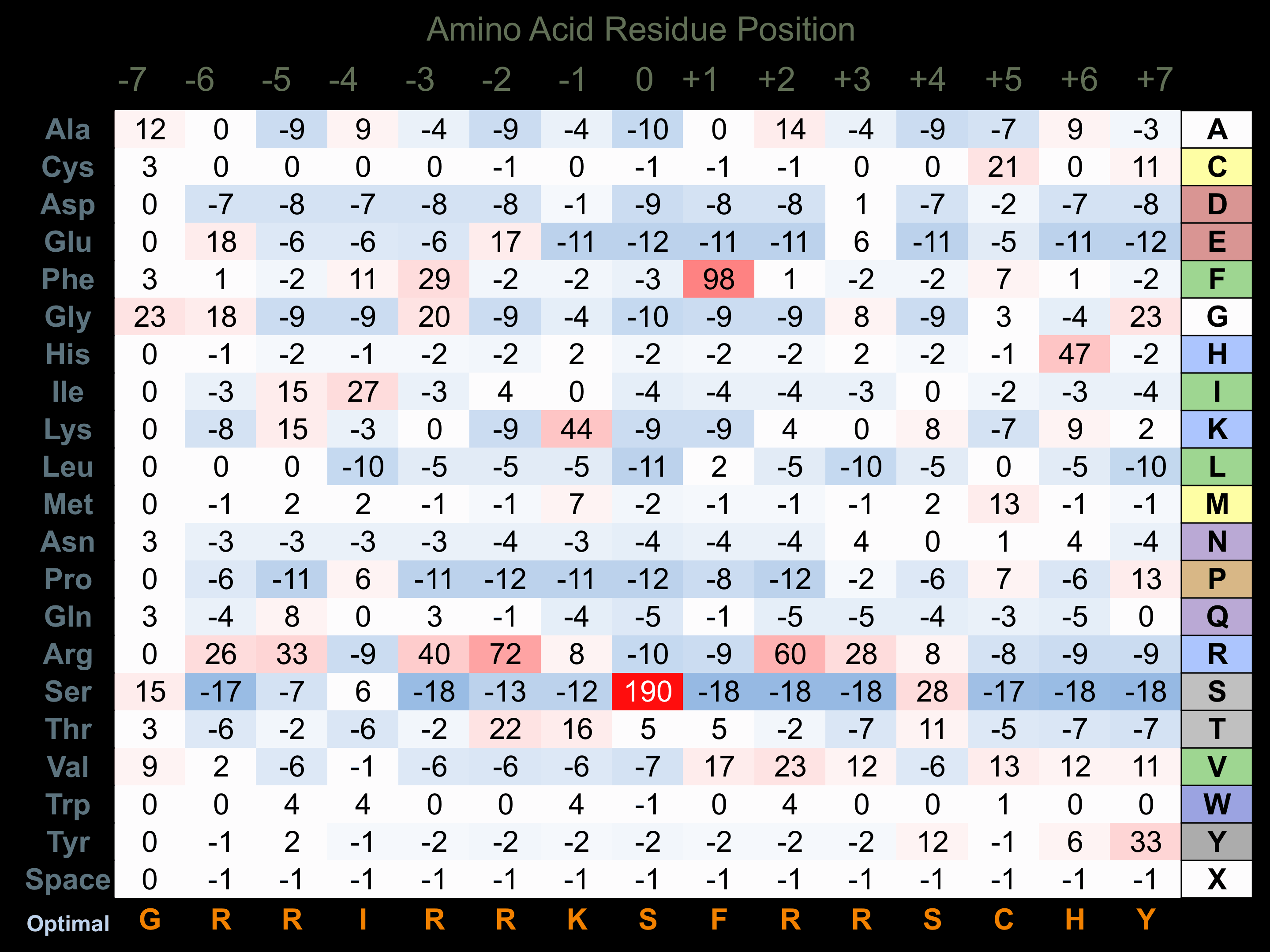
Matrix Type:
Experimentally derived from alignment of 25 known protein substrate phosphosites and 8 peptides phosphorylated by recombinant PKCh in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cardiovascular disorder
Specific Diseases (Non-cancerous):
Stroke, ischemic
Comments:
Mutations in PKCh are associated with susceptibility to ischemic stroke, which is an acute neurologic event leading to death of neural tissue in the brain and loss of cognitive functions. SNPs are involved in cerebral infarction, which is the most common form of stroke.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -56, p<0.002); Cervical cancer (%CFC= +75, p<0.0008); Clear cell renal cell carcinomas (cRCC) (%CFC= +131, p<0.006); Colorectal adenocarcinomas (early onset) (%CFC= +115, p<0.004); Lung adenocarcinomas (%CFC= -55, p<0.0001); Malignant pleural mesotheliomas (MPM) tumours (%CFC= -47, p<0.031); Pituitary adenomas (aldosterone-secreting) (%CFC= +144, p<0.0009); Skin melanomas - malignant (%CFC= -67, p<0.005); Uterine leiomyomas from fibroids (%CFC= -50, p<0.001). The COSMIC website notes an up-regulated expression score for PKCh in diverse human cancers of 277, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 53 for this protein kinase in human cancers was 0.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25218 diverse cancer specimens. This rate is very similar (+ 5% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.36 % in 805 skin cancers tested; 0.35 % in 1184 large intestine cancers tested; 0.27 % in 602 endometrium cancers tested; 0.09 % in 1962 central nervous system cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,501 cancer specimens
Comments:
Only 4 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

