Nomenclature
Short Name:
Akt2
Full Name:
RAC-beta serine-threonine-protein kinase
Alias:
- Akt2
- EC 2.7.11.1
- PKB beta
- RAC-beta serine,threonine protein kinase
- RAC-PK-beta
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
AKT
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
55,769
# Amino Acids:
481
# mRNA Isoforms:
2
mRNA Isoforms:
55,769 Da (481 AA; P31751); 51,083 Da (438 AA; P31751-2)
4D Structure:
Interacts (via PH domain) with MTCP1, TCL1A AND TCL1B. Interacts with TRAF6.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
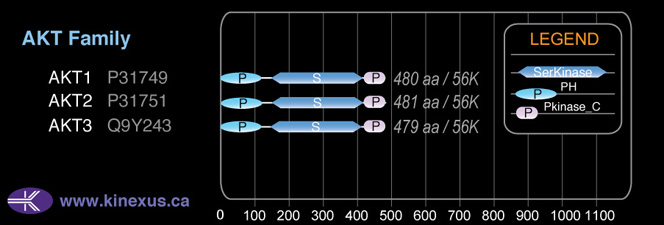
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Tyrosine phosphorylated:
Y38, Y122, Y177, Y178, Y316-, Y327, Y438, Y456, Y475.
Ubiquitinated:
K30, K277, K378.
Acetylated:
K378.
Serine phosphorylated:
S34, S126, S128, S131, S141, S242, S302, S379, S447, S458, S474+, S476, S478.
Threonine phosphorylated:
T81, T306+, T309+, T313, T449, T451.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 71
71
1253
69
1228
 3
3
45
27
45
 24
24
422
16
918
 26
26
455
257
1652
 40
40
704
69
530
 2
2
27
153
91
 10
10
172
91
400
 89
89
1575
80
3328
 14
14
245
24
179
 8
8
146
225
280
 11
11
201
61
507
 29
29
518
305
541
 6
6
113
49
218
 1.2
1.2
21
20
12
 12
12
220
51
398
 4
4
63
44
82
 3
3
53
396
249
 7
7
123
31
171
 8
8
146
204
189
 29
29
517
269
490
 10
10
176
49
389
 12
12
218
54
527
 14
14
252
27
300
 15
15
272
30
970
 16
16
274
49
696
 100
100
1765
156
3380
 5
5
93
52
160
 13
13
226
31
628
 16
16
276
30
867
 17
17
300
84
288
 18
18
311
30
374
 32
32
570
77
1115
 31
31
548
139
1173
 46
46
804
187
725
 8
8
146
100
156
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.8
100 99.6
99.6
99.6
100 -
-
-
99 -
-
-
98 79.4
79.4
89.4
98 -
-
-
- 98.1
98.1
98.8
98 97.7
97.7
98.5
98 -
-
-
- 69.3
69.3
80.1
- 39.7
39.7
55.3
- 87.7
87.7
93.4
- 39.9
39.9
57.2
87 -
-
-
- 49.3
49.3
61.9
64 -
-
-
- 52.7
52.7
69.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
39 -
-
-
40 -
-
-
49 -
-
-
45
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | SH3RF1 - Q7Z6J0 |
| 2 | MTCP1 - P56278 |
| 3 | TCL1A - P56279 |
| 4 | TCL6 - P56846 |
| 5 | TNG2 - P56847 |
| 6 | PDPK1 - O15530 |
| 7 | FSHR - P23945 |
| 8 | GSK3B - P49841 |
| 9 | PRKDC - P78527 |
| 10 | XIAP - P98170 |
| 11 | FOXO4 - P98177 |
| 12 | HSP90AA2 - Q14568 |
| 13 | HSP90AA1 - P07900 |
| 14 | PDPK2 - Q6A1A2 |
| 15 | SETDB1 - Q15047 |
Regulation
Activation:
Phosphorylation of Thr-309 and Ser-474 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ASK1 (MAP3K5) | Q99683 | S83 | ATRGRGSSVGGGSRR | - |
| C2CD5 | Q86YS7 | S197 | EARQRLISLMSGELQ | |
| CLK2 | P49760 | T344 | HHSTIVSTRHYRAPE | - |
| ERa (ESR1) | P03372 | S167 | GGRERLASTNDKGSM | ? |
| Ezrin | P15311 | T566 | QGRDKYKTLRQIRQG | |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| H3.1 | P68431 | S29 | ATKAARKSAPATGGV | + |
| HtrA2 | O43464 | S212 | RVRVRLLSGDTYEAV | |
| IKKa (CHUK) | O15111 | T23 | EMRERLGTGGFGNVC | + |
| PGC-1 | Q9UBK2 | S571 | RMRSRSRSFSRHRSC | |
| PITX2 | Q99697 | T90 | KRQRRQRTHFTSQQL | - |
| PTP1B (PTPN1) | P18031 | S50 | RNRYRDVSPFDHSRI | + |
| QIK (SIK2) | Q9H0K1 | S358 | DGRQRRPSTIAEQTV | + |
| S6 | P62753 | S235 | IAKRRRLSSLRASTS | + |
| S6 | P62753 | S236 | AKRRRLSSLRASTSK | + |
| STXBP4 | Q6ZWJ1 | S99 | RAKLRLESAWEIAFI | |
| XIAP | P98170 | S87 | VGRHRKVSPNCRFIN |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
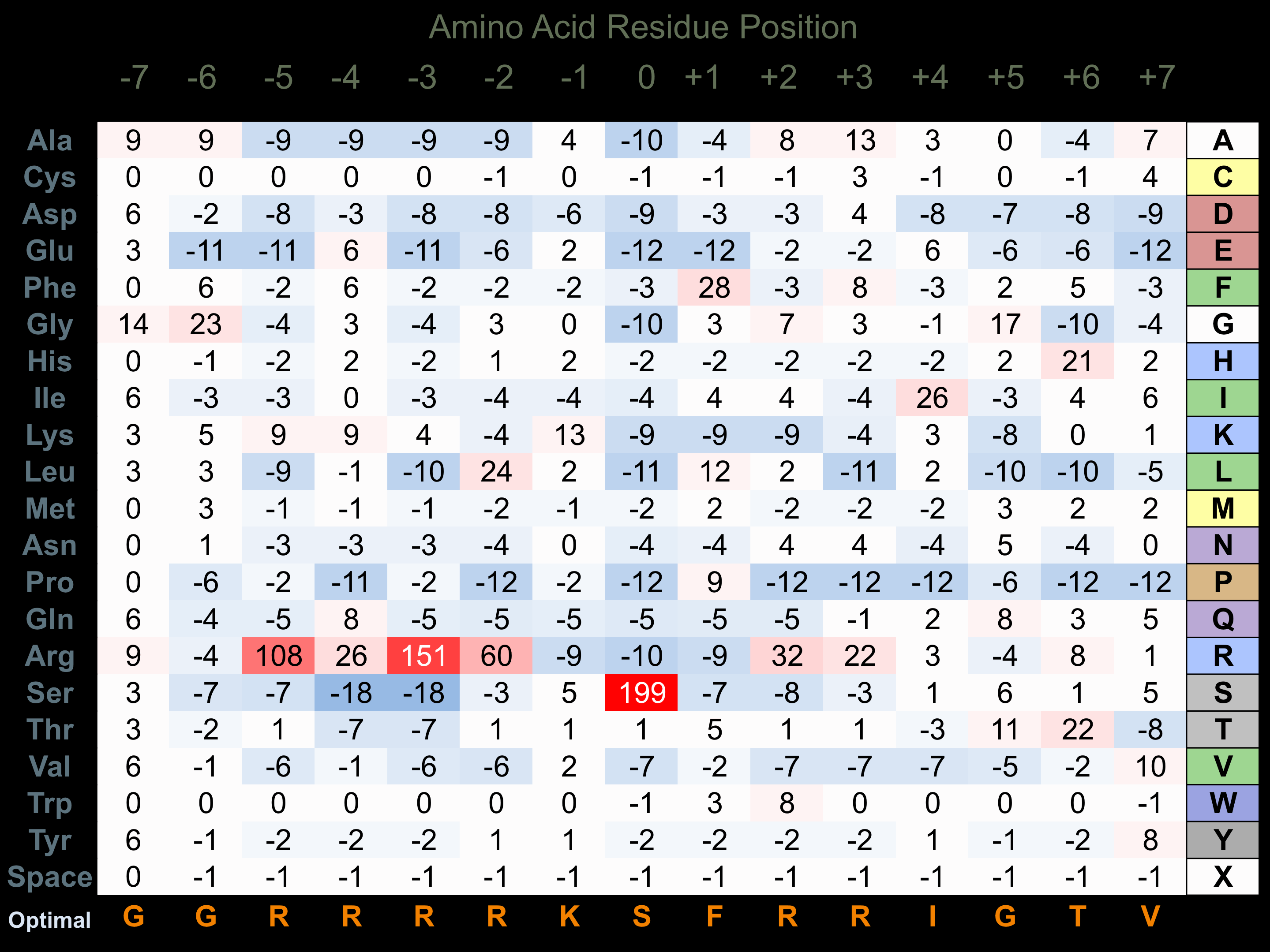
Matrix Type:
Experimentally derived from alignment of 26 known protein substrate phosphosites and 7 peptides phosphorylated by recombinant Akt2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, endocrine disorders
Specific Diseases (Non-cancerous):
Familial partial lipodystrophy due to Akt2 mutations; hypoinsulinemic hypoglycemia with hemihypertrophy (HIHGHH); Insulin resistance; Diabetes mellitus, Type 2; hemihypertrophy; Diabetes mellitus, noninsulin-dependent (NIDDM); hypertension, Insulin resistance-related
Comments:
The R274H mutation in Akt2 is associated with non-insulin-dependent diabetes mellitus, which is a disorder of glucose homeostasis due to lack of recognition of the body's own insulin. R274 in Akt2 normally shields T308 in the activation loop against dephosphorylation, which allows Akt2 to remain catalytically active. Therefore, the R274H mutation is a loss of function mutation. The gain of function E17K mutation is linked to hypoinsulinemic hypoglycemia with hemihypertrophy. It is a disorder with hypoglycemia, low insulin levels, and low serum levels of ketone bodies and branched-chain amino acids, which may lead to reduced consciousness and hypoglycemic seizures. In Akt1, the equivalent E17K mutation leads to plamsa membrane accumulation, catalytic activation and induction of GLUT4 translocation, which induces glucose uptake.
Specific Cancer Types:
Thymoma; ovarian epithelial cancer
Comments:
AKT2 is a known concoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. In many cancers, AKT2 is found to be overexpressed and amplified. It is also linked with invasion, cell migration and induction of angiogenesis. It is important in cell survival regulation, insulin signalling, angiogenesis and tumour formation. Defects in AKT2 contributes to suspectibility to breast cancer and the malignancy of a subset of human ductal pancreatic cancer. AKT2 promotes metastasis of tumour cells, but does not affect the latency of tumour development. It, together with AKT3, has a critical role in glioblastomas. There are no predominent mutations in AKT2 in human cancers, with a relatively dispersed pattern of low level mutations through out the full length of the protein.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain oligodendrogliomas (%CFC= -64, p<0.061); Cervical cancer stage 2B (%CFC= +122, p<0.052); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +1527, p<0.0001); and Ovary adenocarcinomas (%CFC= +81, p<0.003). The COSMIC website notes an up-regulated expression score for AKT2 in diverse human cancers of 798, which is 1.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 212 for this protein kinase in human cancers was 3.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 26371 diverse cancer specimens. This rate is only -1 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.37 % in 1300 large intestine cancers tested; 0.27 % in 910 skin cancers tested; 0.16 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
None >3 in 21,349 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

