Nomenclature
Short Name:
TXK
Full Name:
Tyrosine-protein kinase TXK
Alias:
- BTKL
- EC 2.7.1.112
- RLK
- TKL
- EC 2.7.10.2
- MGC22473
- PSCTK5
- PTK4
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Tec
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
61258
# Amino Acids:
527
# mRNA Isoforms:
1
mRNA Isoforms:
61,258 Da (527 AA; P42681)
4D Structure:
NA
1D Structure:
Subfamily Alignment
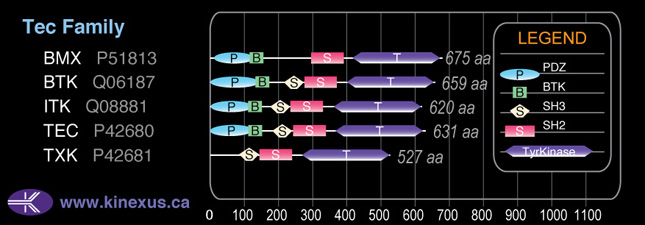
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S307.
Tyrosine phosphorylated:
Y91+, Y420+.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 62
62
778
16
1185
 1.4
1.4
18
12
16
 13
13
169
11
254
 10
10
127
64
262
 17
17
219
14
219
 5
5
58
42
170
 38
38
477
27
527
 31
31
394
41
1281
 17
17
213
10
204
 4
4
55
83
104
 6
6
82
33
136
 24
24
306
152
800
 34
34
436
23
614
 1.3
1.3
16
13
14
 5
5
62
30
191
 0.9
0.9
11
10
8
 1.3
1.3
17
281
56
 16
16
204
21
272
 3
3
37
66
70
 18
18
230
56
251
 4
4
45
28
158
 10
10
125
32
168
 4
4
49
28
71
 5
5
68
21
93
 12
12
155
31
198
 45
45
570
40
1264
 13
13
163
26
229
 3
3
37
19
31
 6
6
78
21
170
 6
6
79
14
51
 100
100
1265
18
724
 36
36
451
15
389
 7
7
93
55
235
 40
40
507
36
600
 2
2
27
22
25
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 86.7
86.7
87.1
99 97
97
98.3
97 -
-
-
89 -
-
-
92 79
79
84.4
88 -
-
-
- 83.5
83.5
92.4
83 36.2
36.2
56.2
82 -
-
-
- 51.1
51.1
67.2
- 45.2
45.2
59.2
66 36.6
36.6
52.6
- -
-
-
62 -
-
-
- 33.7
33.7
45.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
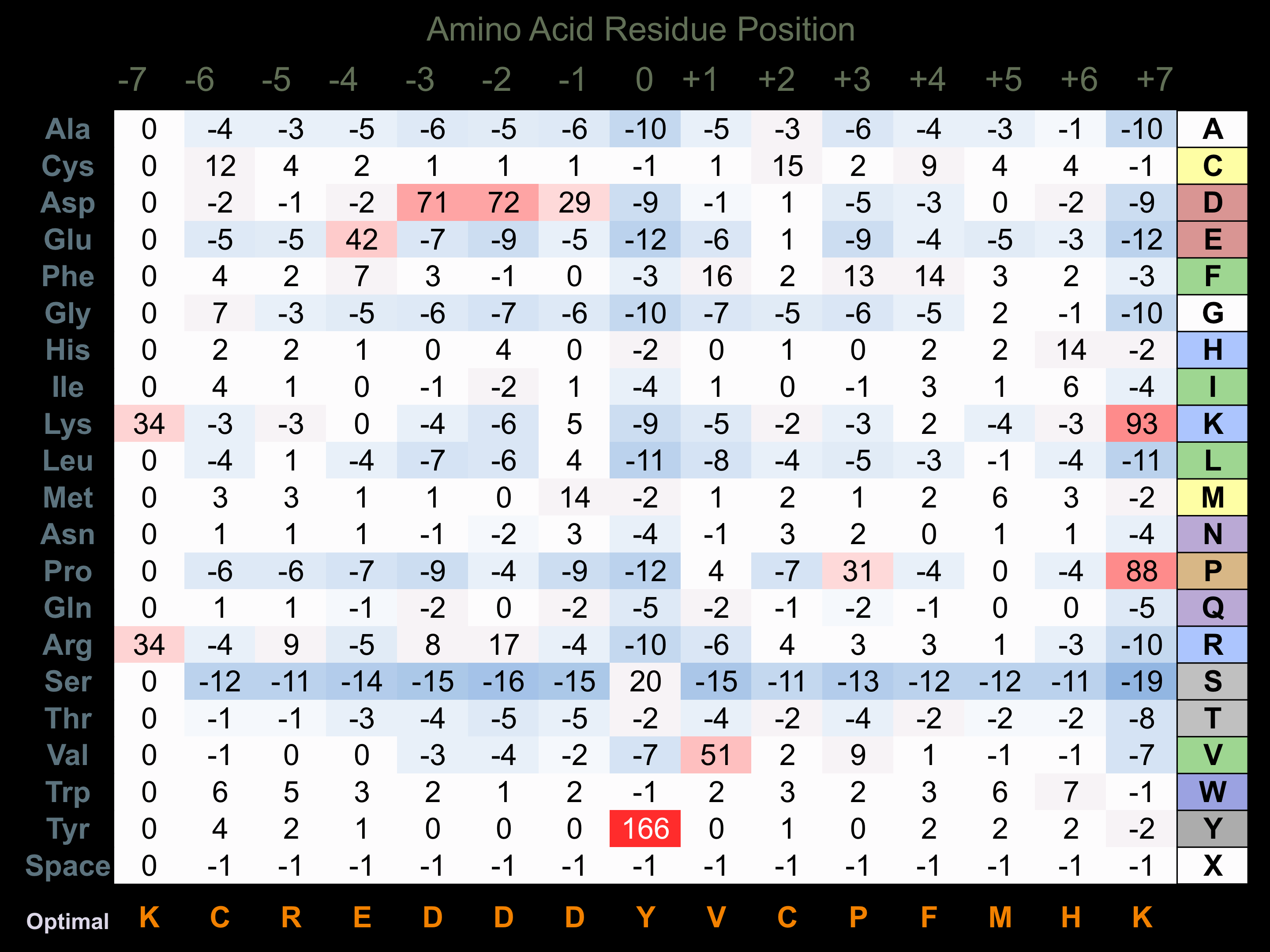
Matrix Type:
Experimentally derived from alignment of 2 known protein substrate phosphosites and 84 peptides phosphorylated by recombinant TXK in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Immune disorder
Specific Diseases (Non-cancerous):
Behcet's disease
Comments:
Behcet's disease (BD), also known as silk road disease, is a rare immune disease characterized by small blood vessel vasculitis, mucous membrane ulceration, and ocular defects. Affected systems include the gastrointestinal tract, pulmonary, musculoskeletal, cardiovascular, and nervous system. This disease is often fatal due to aneurysms or severe neurological deficits. Common symptoms include sores and swelling on the skin, genitals, mouth, and parts of the eye, which can be complicated by meningitis, blood clots, digestive tract inflammation, and blindness. Behcet's disease is more common in Japan, Turkey, and Israel. In animal studies, mice heterozygous for a loss-of-function mutation of the TXK gene did not display any phenotypic effects, whereas mice homozygous for the loss-of-function mutation displayed significantly reduced numbers of mature T-cells, particularly CD4+ cells. Additionally, the small number of T-cells present displayed abnormal cell proliferation, cytokine synthesis, and apoptosis in response to T-cell receptor activation, indicating dysfunctional activity. The TXK protein has been previously shown to function as a Th1 specific transcription factor involved in the effector function of Th1 cells. Th1 cells are implicated in BD as they aberrantly accumulate in the skin and intestinal lesions of BD patients. Interestingly, these Th1 cells have been shown to display elevated levels of TXK expression, therefore TXK may have a role in the pathogenesis of the disease.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24956 diverse cancer specimens. This rate is only 14 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.52 % in 805 skin cancers tested; 0.42 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,197 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

