Nomenclature
Short Name:
FYN
Full Name:
Proto-oncogene tyrosine-protein kinase FYN
Alias:
- EC 2.7.10.2
- P59-Fyn
- SLK
- SYN
- FYN
- FYN oncogene related to SRC, FGR, YES
- Kinase Fyn
- MGC45350
- P59-FYN
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Src
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
60762
# Amino Acids:
537
# mRNA Isoforms:
3
mRNA Isoforms:
60,762 Da (537 AA; P06241); 60,141 Da (534 AA; P06241-2); 54,513 Da (482 AA; P06241-3)
4D Structure:
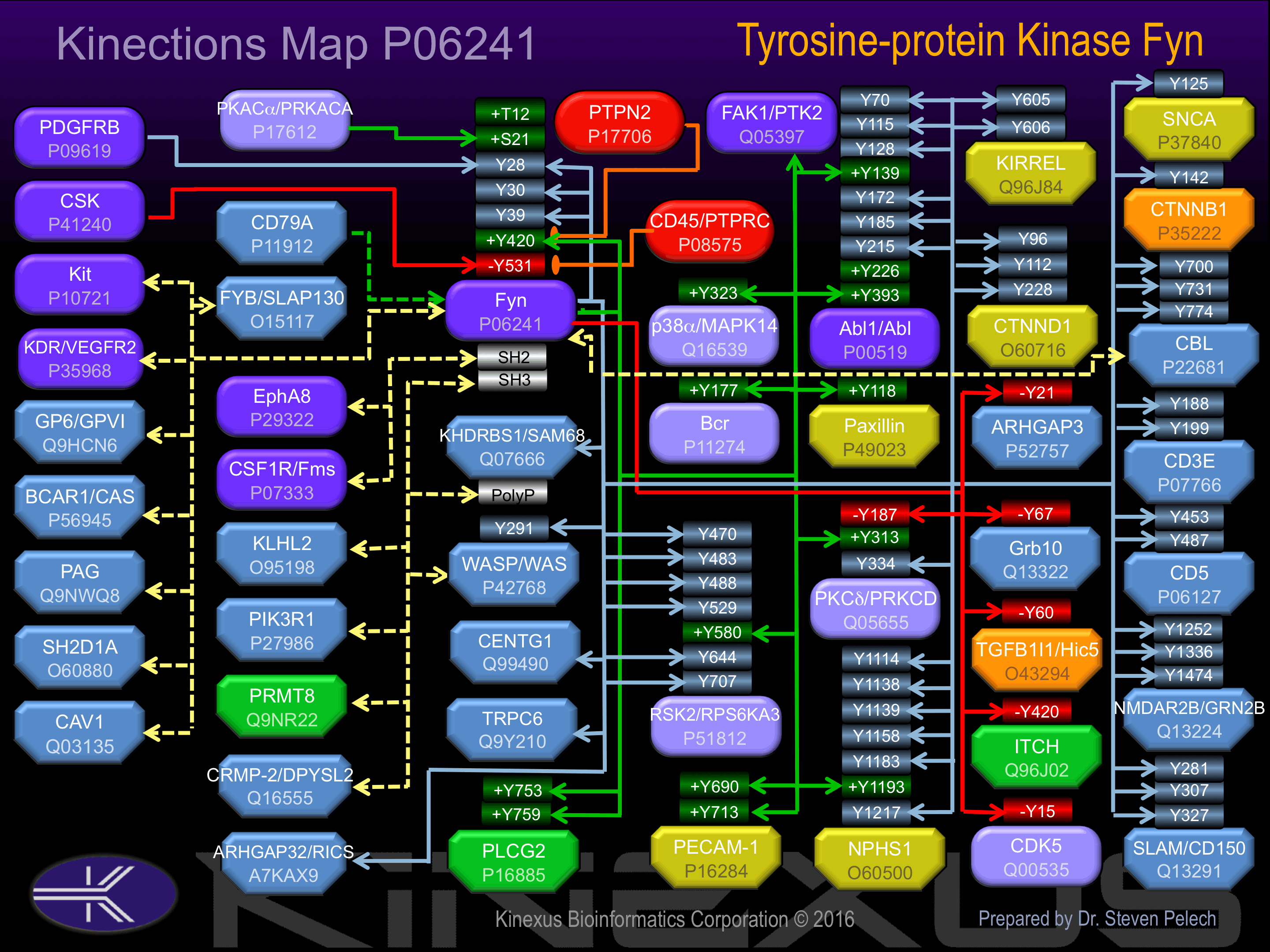
Interacts (via its SH3 domain) with PIK3R1 and PRMT8. Interacts with FYB, PAG1, and SH2D1A. Interacts with CD79A (tyrosine-phosphorylated form); the interaction increases FYN activity By similarity. Interacts with TOM1L1 (phosphorylated form) By similarity. Interacts with SH2D1A and SLAMF1. Interacts (via its SH3 domain) with HEV ORF3 protein. Interacts with ITCH; the interaction phosphorylates ITCH and negatively regulates its activity.
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S21+, S25, S26, S178, S186, S188, S257, S307, S349, S393.
Threonine phosphorylated:
T12, T181, T216, T217, T289, T294, T300, T421, T525, T527.
Tyrosine phosphorylated:
Y28, Y30, Y39, Y91, Y185, Y213, Y214, Y339, Y420+, Y440, Y483, Y531-.
Ubiquitinated:
K182, K405, K431.
Myristoylated:
G2.
Palmitoylated:
C3 (predicted), C6 (predicted).
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 26
26
991
57
767
 2
2
75
25
72
 11
11
408
13
348
 18
18
673
170
626
 24
24
911
38
604
 3
3
109
133
139
 9
9
333
65
496
 11
11
416
72
530
 17
17
630
24
481
 6
6
221
149
200
 5
5
184
52
218
 20
20
767
328
732
 11
11
426
46
439
 3
3
122
23
130
 2
2
78
43
95
 2
2
81
29
100
 8
8
291
397
2646
 2
2
86
28
93
 2
2
85
139
86
 21
21
796
185
689
 17
17
627
40
680
 12
12
437
46
512
 8
8
289
24
222
 4
4
136
29
133
 11
11
400
40
439
 17
17
654
104
682
 6
6
245
49
243
 3
3
121
29
113
 7
7
268
30
314
 2
2
61
42
62
 36
36
1361
42
578
 100
100
3794
56
9386
 14
14
526
97
884
 19
19
738
109
661
 3
3
128
79
216
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.9
98.9
98.9
95 99.6
99.6
99.8
95 -
-
-
95 -
-
-
98 99.8
99.8
99.8
95 -
-
-
- 99.1
99.1
99.6
99 99.6
99.6
99.6
95 -
-
-
- 70.1
70.1
73.3
- 92.4
92.4
94.4
97 96.8
96.8
98.1
94 90.6
90.6
95.2
87 -
-
-
- 54.6
54.6
68.2
- -
-
-
- -
-
-
54.5 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CBL - P22681 |
| 2 | FYB - O15117 |
| 3 | FCER2 - P06734 |
| 4 | BCAR1 - P56945 |
| 5 | CTLA4 - P16410 |
| 6 | IL7R - P16871 |
| 7 | CSK - P41240 |
| 8 | GP6 - Q9HCN6 |
| 9 | ITK - Q08881 |
| 10 | ARHGAP32 - A7KAX9 |
| 11 | SLAMF1 - Q13291 |
| 12 | PIK3R1 - P27986 |
| 13 | GRIN2B - Q13224 |
| 14 | CD36 - P16671 |
| 15 | DLG4 - P78352 |
Regulation
Activation:
NA
Inhibition:
Phosphorylation of Tyr-531 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Abl1 | P00519 | Y115 | QGWVPSNYITPVNSL | |
| Abl1 | P00519 | Y128 | SLEKHSWYHGPVSRN | |
| Abl1 | P00519 | Y139 | VSRNAAEYLLSSGIN | + |
| Abl1 | P00519 | Y172 | LRYEGRVYHYRINTA | |
| Abl1 | P00519 | Y185 | TASDGKLYVSSESRF | |
| Abl1 | P00519 | Y215 | GLITTLHYPAPKRNK | |
| Abl1 | P00519 | Y226 | KRNKPTVYGVSPNYD | + |
| Abl1 | P00519 | Y393 | RLMTGDTYTAHAGAK | + |
| Abl1 | P00519 | Y70 | PNLFVALYDFVASGD | + |
| Abl1 iso2 | P00519-2 | Y134 | QGWVPSNYITPVNSL | |
| Abl1 iso2 | P00519-2 | Y147 | SLEKHSWYHGPVSRN | |
| Abl1 iso2 | P00519-2 | Y158 | VSRNAAEYLLSSGIN | + |
| Abl1 iso2 | P00519-2 | Y191 | LRYEGRVYHYRINTA | |
| Abl1 iso2 | P00519-2 | Y204 | TASDGKLYVSSESRF | |
| Abl1 iso2 | P00519-2 | Y234 | GLITTLHYPAPKRNK | |
| Abl1 iso2 | P00519-2 | Y245 | KRNKPTVYGVSPNYD | + |
| Abl1 iso2 | P00519-2 | Y412 | RLMTGDTYTAHAGAK | + |
| Abl1 iso2 | P00519-2 | Y89 | PNLFVALYDFVASGD | + |
| ADD2 | P35612 | Y489 | PNQFVPLYTDPQEVL | |
| APP | P05067 | Y757 | SKMQQNGYENPTYKF | |
| ARHGAP3 | P52757 | Y21 | VSSDAEEYQPPIWKS | - |
| Bcr | P11274 | Y177 | ADAEKPFYVNVEFHH | + |
| Calponin 1 | P51911 | Y182 | SQQGMTAYGTRRHLY | |
| Calponin 1 | P51911 | Y261 | SQRGMTVYGLPRQVY | |
| Calponin 3 | Q15417 | Y261 | SQKGMSVYGLGRQVY | |
| Caveolin 1 (CAV1) | Q03135 | Y14 | VDSEGHLYTVPIREQ | |
| Cbl | P22681 | Y700 | EGEEDTEYMTPSSRP | |
| Cbl | P22681 | Y731 | QQIDSCTYEAMYNIQ | |
| Cbl | P22681 | Y774 | SENEDDGYDVPKPPV | |
| CD3E | P07766 | Y188 | PPVPNPDYEPIRKGQ | |
| CD3E | P07766 | Y199 | RKGQRDLYSGLNQRR | |
| CD5 | P06127 | Y453 | ASHVDNEYSQPPRNS | |
| CD5 | P06127 | Y487 | DNSSDSDYDLHGAQR | |
| CDK5 | Q00535 | Y15 | EKIGEGTYGTVFKAK | - |
| CENTG1 iso2 | Q99490 | Y682 | ESWIRAKYEQLLFLA | |
| CENTG1 iso2 | Q99490 | Y774 | QGRTALFYARQAGSQ | |
| CLIC5 | Q9NZA1 | Y33 | EENESPHYDDVHEYL | |
| CTLA-4 | P16410 | Y201 | SPLTTGVYVKMPPTE | |
| CTLA-4 | P16410 | Y218 | CEKQFQPYFIPIN__ | |
| CTNNB1 | P35222 | Y142 | AVVNLINYQDDAELA | |
| CTNND1 | O60716 | Y112 | PGQIVETYTEEDPEG | |
| CTNND1 | O60716 | Y228 | YPGGSDNYGSLSRVT | |
| CTNND1 | O60716 | Y96 | QDHSHLLYSTIPRMQ | |
| DCC | P43146 | Y1261 | PTLESAQYPGILPSP | |
| DCC | P43146 | Y1420 | TEDSANVYEQDDLSE | |
| Desmoplakin 3 | P14923 | Y132 | AIVHLINYQDDAELV | |
| Desmoplakin 3 | P14923 | Y548 | AAGTQQPYTDGVRME | |
| DLG4 (PSD-95) | P78352 | Y523 | REDSVLSYETVTQME | |
| FcGR2A | P12318 | Y288 | YETADGGYMTLNPRA | |
| FcGR2A | P12318 | Y304 | TDDDKNIYLTLPPND | |
| FcGR2B | P31994 | Y292 | GAENTITYSLLMHPD | |
| FcGR2C | P31995 | Y310 | TDDDKNIYLTLPPND | |
| FLOT1 | O75955 | Y160 | DIHDDQDYLHSLGKA | |
| FLOT2 | Q14254 | Y114 | DVYDKVDYLSSLGKT | |
| Fyn | P06241 | Y28 | SLNQSSGYRYGTDPT | |
| Fyn | P06241 | Y30 | NQSSGYRYGTDPTPQ | |
| Fyn | P06241 | Y39 | TDPTPQHYPSFGVTS | |
| Fyn | P06241 | Y420 | RLIEDNEYTARQGAK | + |
| Grb10 | Q13322 | Y67 | NASLESLYSACSMQS | - |
| H3.1 | P68431 | S10 | TKQTARKSTGGKAPR | |
| H3.2 | P84228 | S11 | TKQTARKSTGGKAPR | |
| IP3R1 (IP3 receptor) | Q14643 | Y353 | NAQEKMVYSLVSVPE | |
| IREM1 | Q8TDQ1 | Y236 | VDQVEVEYVTMASLP | |
| IREM1 | Q8TDQ1 | Y263 | AEDQEPTYCNMGHLS | |
| IRS1 | P35568 | Y1179 | GLENGLNYIDLDLVK | |
| IRS1 | P35568 | Y896 | EPKSPGEYVNIEFGS | ? |
| ITCH | Q96J02 | Y420 | QFNQRFIYGNQDLFA | - |
| KIRREL | Q96J84 | Y605 | MKDPTNGYYNVRAHE | |
| KIRREL | Q96J84 | Y606 | KDPTNGYYNVRAHED | |
| LMR1 (AATK) | Q6ZMQ8 | Y73 | ENAEGDEYAADLAQG | |
| LMR1 (AATK) | Q6ZMQ8 | Y93 | AQNGPDVYVLPLTEV | |
| MAP2 | P11137 | Y67 | EHGSQGTYSNTKENG | |
| MAP2 iso3 | P11137 | Y50 | RSANGFPYREDEEGA | |
| NFE2L2 | Q16236 | Y576 | RDEDGKPYSPSEYSL | |
| NMDAR2B (GRIN2B) | Q13224 | Y1252 | CKKAGNLYDISEDNS | |
| NMDAR2B (GRIN2B) | Q13224 | Y1336 | RFMDGSPYAHMFEMS | |
| NMDAR2B (GRIN2B) | Q13224 | Y1474 | GSSNGHVYEKLSSIE | ? |
| NMT1 | P30419 | Y117 | EEASKRSYQFWDTQP | |
| NMT1 | P30419 | Y180 | YTLLNENYVEDDDNM | + |
| Nogo | Q9NQC3 | Y694 | LQETEAPYISIACDL | |
| NPHS1 | O60500 | Y1114 | EDRVRNEYEESQWTG | |
| NPHS1 | O60500 | Y1138 | STTEAEPYYRSLRDF | |
| NPHS1 | O60500 | Y1139 | TTEAEPYYRSLRDFS | |
| NPHS1 | O60500 | Y1158 | PTQEEVSYSRGFTGE | |
| NPHS1 | O60500 | Y1183 | YDEVERTYPPSGAWG | |
| NPHS1 | O60500 | Y1193 | SGAWGPLYDEVQMGP | + |
| NPHS1 | O60500 | Y1217 | YQDPRGIYDQVAGDL | |
| p38a MAPK (MAPK14) | Q16539 | Y323 | DEPVADPYDQSFESR | + |
| Paxillin (PXN) | P49023 | Y118 | VGEEEHVYSFPNKQK | + |
| PECAM-1 | P16284 | Y690 | PLNSDVQYTEVQVSS | + |
| PECAM-1 | P16284 | Y713 | KKDTETVYSEVRKAV | + |
| PKCd (PRKCD) | Q05655 | Y187 | WGLNKQGYKCRQCNA | - |
| PKCd (PRKCD) | Q05655 | Y313 | SSEPVGIYQGFEKKT | + |
| PKCd (PRKCD) | Q05655 | Y334 | MQDNSGTYGKIWEGS | ? |
| PLCG2 | P16885 | Y753 | ERDINSLYDVSRMYV | + |
| PLCG2 | P16885 | Y759 | LYDVSRMYVDPSEIN | + |
| PLD2 | O14939 | T11 | TPESLFPTGDELDSS | |
| PLD2 | O14939 | Y165 | HAASKQKYLENYLNR | |
| PLD2 | O14939 | Y470 | GRWDDLHYRLTDLGD | |
| PROM1 | O43490 | Y828 | RMDSEDVYDDVETIP | |
| PROM1 | O43490 | Y852 | GYHKDHVYGIHNPVM | |
| PSD-93 | Q15700 | Y348 | TRPPEPVYSTVNKLC | |
| RA70 | O75563 | Y232 | EEEKEETYDDIDGFD | |
| RA70 | O75563 | Y261 | QPIDDEIYEELPEEE | |
| RA70 | O75563 | Y295 | TRRKGVDYASYYQGL | |
| RA70 | O75563 | Y298 | KGVDYASYYQGLWDC | |
| RSK2 (RPS6KA3) | P51812 | Y470 | EIEILLRYGQHPNII | |
| RSK2 (RPS6KA3) | P51812 | Y483 | IITLKDVYDDGKYVY | |
| RSK2 (RPS6KA3) | P51812 | Y488 | DVYDDGKYVYVVTEL | |
| RSK2 (RPS6KA3) | P51812 | Y529 | TITKTVEYLHAQGVV | ? |
| RSK2 (RPS6KA3) | P51812 | Y580 | GLLMTPCYTANFVAP | + |
| RSK2 (RPS6KA3) | P51812 | Y644 | KFSLSGGYWNSVSDT | |
| RSK2 (RPS6KA3) | P51812 | Y707 | KGAMAATYSALNRNQ | |
| SCN5A | Q14524 | Y1494 | MTEEQKKYYNAMKKL | |
| SCN5A | Q14524 | Y1495 | TEEQKKYYNAMKKLG | |
| SIGLEC10 | Q96LC7 | Y667 | ESQEELHYATLNFPG | |
| SLAM (CD150) | Q13291 | Y281 | EKKSLTIYAQVQKPG | |
| SLAM (CD150) | Q13291 | Y307 | QDPCTTIYVAATEPV | |
| SLAM (CD150) | Q13291 | Y327 | ETNSITVYASVTLPE | |
| SNCA | P37840 | Y125 | VDPDNEAYEMPSEEG | |
| SNX26 | O14559 | Y406 | PLLTYQLYGKFSEAM | |
| Srcasm | O75674 | Y442 | DLQPPNYYEVMEFDP | |
| Srcasm | O75674 | Y460 | AVTTEAIYEEIDAHQ | |
| Tau iso5 (Tau-C) | P10636-5 | Y18 | MEDHAGTYGLGDRKD | |
| TGFB1I1 (Hic-5) | O43294 | Y60 | SGDKDHLYSTVCKPR | - |
| TXK | P42681 | Y420 | RYVLDDEYVSSFGAK | + |
| WASP | P42768 | Y291 | AETSKLIYDFIEDQG | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
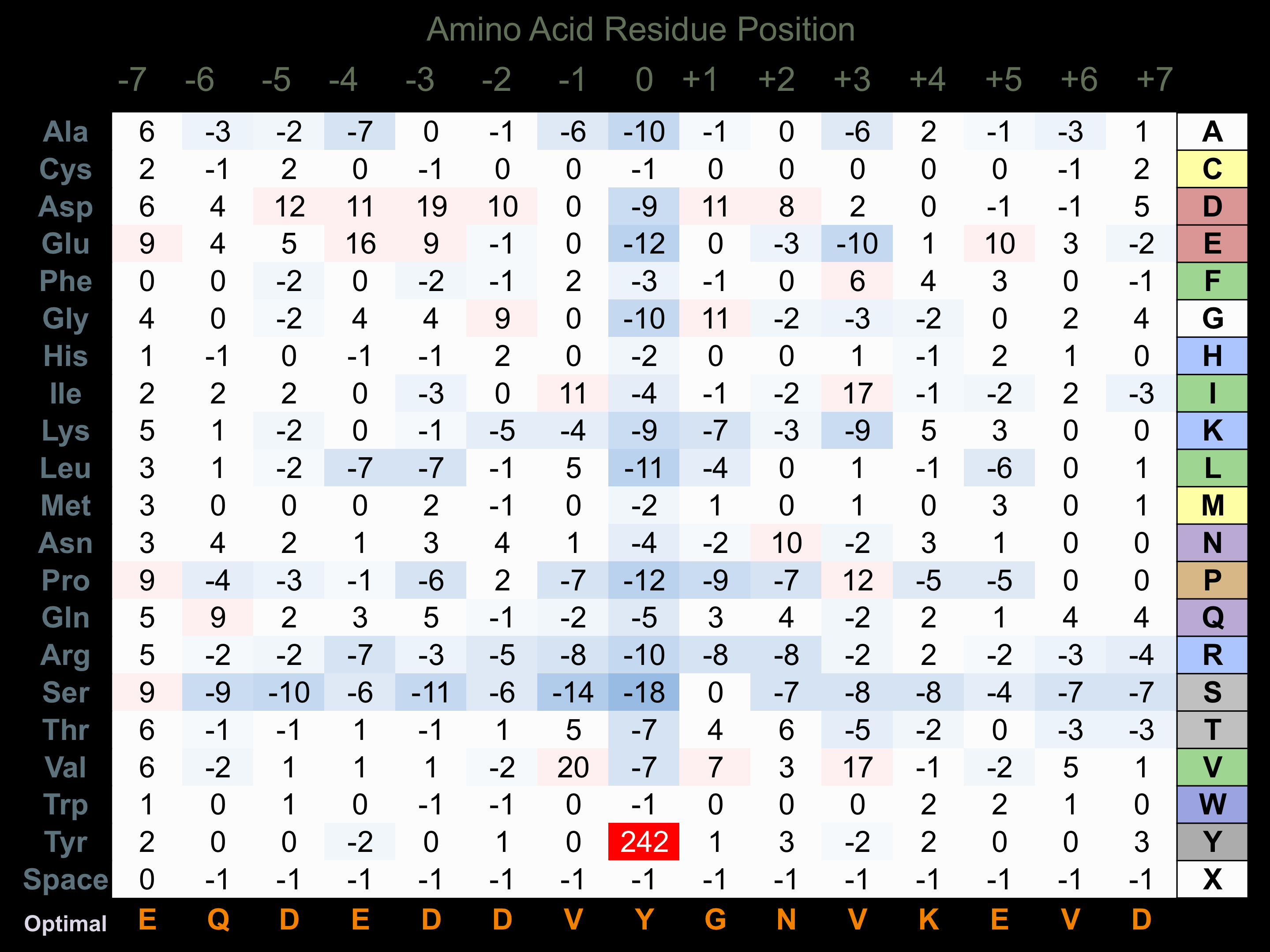
Matrix Type:
Experimentally derived from alignment of 165 known protein substrate phosphosites and 10 peptides phosphorylated by recombinant Fyn in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Estrogen-receptor negative breast cancer
Comments:
FYN may be an oncoprotein (OP) based on its similarity to other Src family kinases that are known oncoproteins. The active form of the protein kinase normally acts to promote tumour cell proliferation. Fyn has been linked with estrogen-receptor negative breast cancer, which is related to breast and colon cancer, and it may affect breast, lymph node, or lung tissues.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +116, p<0.036); Classical Hodgkin lymphomas (%CFC= +48, p<0.038); Colon mucosal cell adenomas (%CFC= -47, p<0.0001); Colorectal adenocarcinomas (early onset) (%CFC= +154, p<(0.0003); Large B-cell lymphomas (%CFC= +51, p<0.06); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +62, p<0.026); Oral squamous cell carcinomas (OSCC) (%CFC= +54, p<0.076); Papillary thyroid carcinomas (PTC) (%CFC= +66, p<0.022); Skin melanomas - malignant (%CFC= +100, p<0.001); T-cell prolymphocytic leukemia (%CFC= -60, p<0.002); and Uterine fibroids (%CFC= +66, p<0.015). The COSMIC website notes an up-regulated expression score for FYN in diverse human cancers of 270, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 18 for this protein kinase in human cancers was 0.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25627 diverse cancer specimens. This rate is only 24 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.56 % in 1093 large intestine cancers tested; 0.42 % in 805 skin cancers tested; 0.41 % in 589 stomach cancers tested; 0.17 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R206C (6).
Comments:
Only 2 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.