Nomenclature
Short Name:
LYN
Full Name:
Tyrosine-protein kinase Lyn
Alias:
- EC 2.7.10.2
- FLJ13161
- LYN
- V-yes-1 Yamaguchi sarcoma viral related oncogene
- FLJ26625
- JTK8
- Kinase Lyn
- KPM
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Src
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
58574
# Amino Acids:
512
# mRNA Isoforms:
2
mRNA Isoforms:
58,574 Da (512 AA; P07948); 56,033 Da (491 AA; P07948-2)
4D Structure:
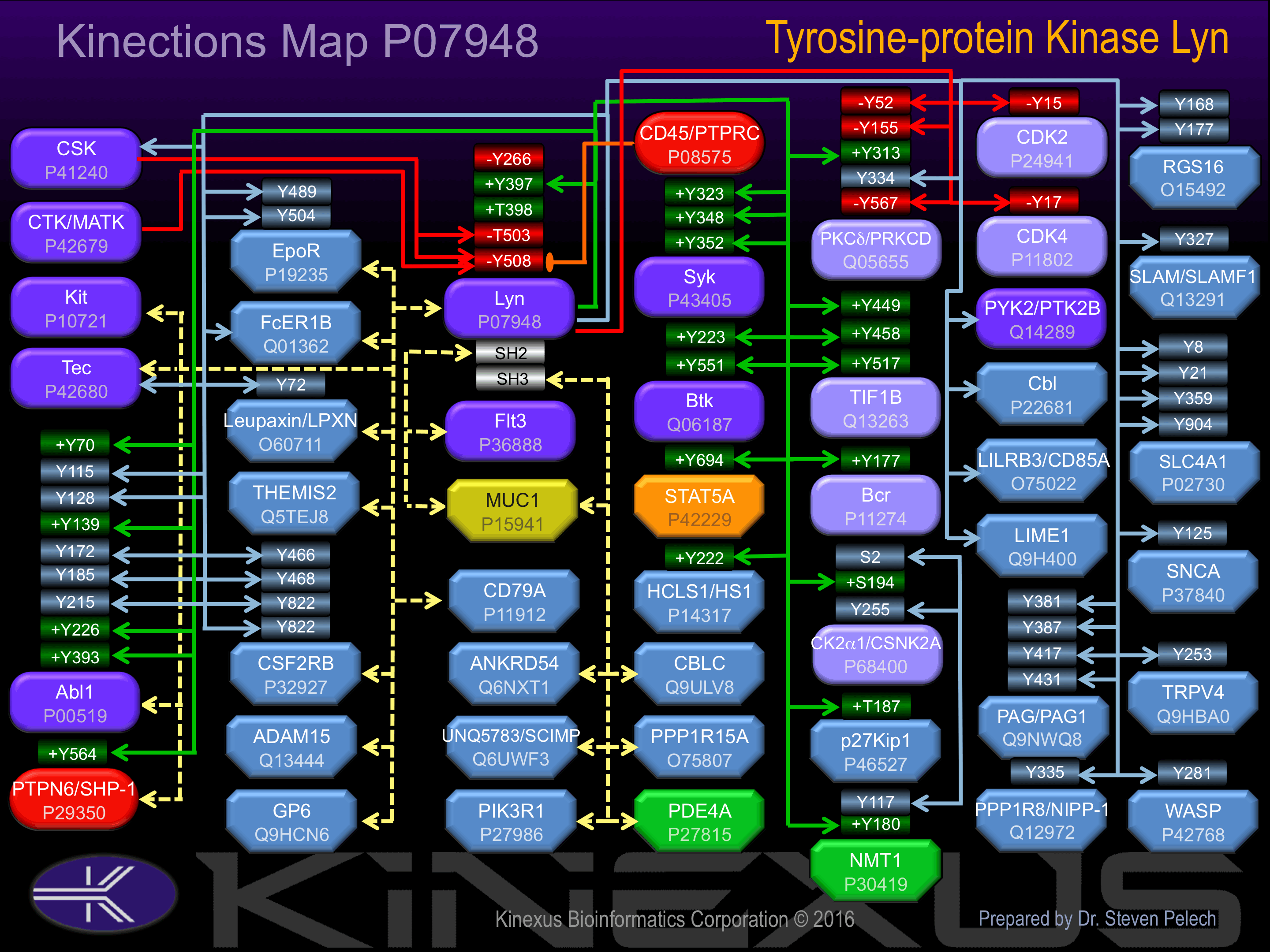
Interacts with phosphorylated LIME1 and with CD79A upon BCR activation. Interacts with Epstein-Barr virus LMP2A. Interacts with TGFB1I1. Interaction, via the SH2 and SH3, domains with MUC1 is stimulated by IL7 and, the subsequent phosphorylation increases the binding between MUC1 and CTNNB1/beta-catenin. Interacts with PPP1R15A and PDE4A via the SH3 domain. Interacts with Herpesvirus saimiri tyrosine kinase interacting protein (Tip). Interacts with ADAM15. Interacts with NDFIP2 and more weakly with NDFIP1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Myristoylated:
G2.
Palmitoylated:
C3.
Serine phosphorylated:
S11, S13, S115, S149, S158, S164, S228, S246, S269, S283, S326.
Threonine phosphorylated:
T58, T135, T276, T281, T296, T319, T398+, T438, T448, T502-, T503-, T504.
Tyrosine phosphorylated:
Y32, Y117, Y193, Y194, Y265, Y266-, Y306, Y316, Y321, Y397+, Y439, Y460, Y473, Y501, Y508-.
Ubiquitinated:
K9, K20, K59, K477.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 46
46
1143
42
1267
 4
4
95
23
45
 3
3
86
14
56
 18
18
442
147
928
 34
34
847
36
660
 5
5
126
111
89
 17
17
411
51
558
 44
44
1079
69
2719
 40
40
988
24
632
 3
3
78
141
50
 4
4
93
50
70
 31
31
768
282
695
 10
10
246
47
111
 2
2
46
21
29
 4
4
90
41
88
 4
4
99
24
41
 4
4
94
366
747
 4
4
101
28
57
 2
2
44
134
26
 22
22
538
162
552
 4
4
87
39
45
 21
21
513
45
335
 6
6
153
25
98
 2
2
39
29
35
 3
3
79
39
55
 23
23
574
89
657
 15
15
372
50
164
 8
8
192
29
83
 3
3
64
29
35
 4
4
106
42
82
 24
24
590
30
599
 39
39
972
51
754
 100
100
2474
88
2512
 67
67
1648
83
600
 20
20
498
48
993
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 57.3
57.3
68.5
100 80
80
81.5
99 -
-
-
97 -
-
-
98 97
97
99.2
97 -
-
-
- 96
96
98.8
96 96
96
98.8
96 -
-
-
- 93.1
93.1
96.6
- 63
63
77.7
88 56.7
56.7
72
86 76.3
76.3
86.9
77.5 -
-
-
- 51.4
51.4
69.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
Phosphorylation of Thr-503 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Abl1 | P00519 | Y115 | QGWVPSNYITPVNSL | |
| Abl1 | P00519 | Y128 | SLEKHSWYHGPVSRN | |
| Abl1 | P00519 | Y139 | VSRNAAEYLLSSGIN | + |
| Abl1 | P00519 | Y172 | LRYEGRVYHYRINTA | |
| Abl1 | P00519 | Y185 | TASDGKLYVSSESRF | |
| Abl1 | P00519 | Y215 | GLITTLHYPAPKRNK | |
| Abl1 | P00519 | Y226 | KRNKPTVYGVSPNYD | + |
| Abl1 | P00519 | Y393 | RLMTGDTYTAHAGAK | + |
| Abl1 | P00519 | Y70 | PNLFVALYDFVASGD | + |
| Abl1 iso2 | P00519-2 | Y134 | QGWVPSNYITPVNSL | |
| Abl1 iso2 | P00519-2 | Y147 | SLEKHSWYHGPVSRN | |
| Abl1 iso2 | P00519-2 | Y158 | VSRNAAEYLLSSGIN | + |
| Abl1 iso2 | P00519-2 | Y191 | LRYEGRVYHYRINTA | |
| Abl1 iso2 | P00519-2 | Y204 | TASDGKLYVSSESRF | |
| Abl1 iso2 | P00519-2 | Y234 | GLITTLHYPAPKRNK | |
| Abl1 iso2 | P00519-2 | Y245 | KRNKPTVYGVSPNYD | + |
| Abl1 iso2 | P00519-2 | Y412 | RLMTGDTYTAHAGAK | + |
| Abl1 iso2 | P00519-2 | Y89 | PNLFVALYDFVASGD | + |
| Bcr | P11274 | Y177 | ADAEKPFYVNVEFHH | + |
| Btk | Q06187 | Y223 | LKKVVALYDYMPMNA | + |
| Btk | Q06187 | Y551 | RYVLDDEYTSSVGSK | + |
| Caspase 8 | Q14790 | Y380 | TDSEEQPYLEMDLSS | |
| Caspase 8 | Q14790 | Y448 | TILTEVNYEVSNKDD | |
| CD19 | P15391 | Y500 | TSLGSQSYEDMRGIL | |
| CD19 | P15391 | Y531 | HEEDADSYENMDNPD | |
| CDK2 | P24941 | Y15 | EKIGEGTYGVVYKAR | - |
| CDK4 | P11802 | Y17 | AEIGVGAYGTVYKAR | - |
| CK2a1 (CSNK2A1) | P68400 | S194 | EYNVRVASRYFKGPE | + |
| CK2a1 (CSNK2A1) | P68400 | S2 | ______MSGPVPSRA | |
| CK2a1 (CSNK2A1) | P68400 | Y255 | VLGTEDLYDYIDKYN | |
| CSF2RB | P32927 | Y466 | ALRFCGIYGYRLRRK | |
| CSF2RB | P32927 | Y468 | RFCGIYGYRLRRKWE | |
| CSF2RB | P32927 | Y822 | VLQQVGDYCFLPGLG | ? |
| CSF2RB | P32927 | Y882 | KALKQQDYLSLPPWE | ? |
| DAPP1 | Q9UN19 | Y139 | KVEEPSIYESVRVHT | |
| DOK2 | O60496 | Y271 | LPRPDSPYSRPHDSL | |
| DOK2 | O60496 | Y299 | PRGQEGEYAVPFDAV | |
| DOK2 | O60496 | Y345 | PPRPDHIYDEPEGVA | |
| EpoR | P19235 | Y489 | DGPYSNPYENSLIPA | ? |
| EpoR | P19235 | Y504 | AEPLPPSYVACS___ | ? |
| FcGR2A | P12318 | Y304 | TDDDKNIYLTLPPND | |
| FcGR2B | P31994 | Y292 | GAENTITYSLLMHPD | |
| FcGR2C | P31995 | Y310 | TDDDKNIYLTLPPND | |
| GCET2 | Q8N6F7 | Y107 | GNSAEEYYENVPCKA | |
| GCET2 | Q8N6F7 | Y128 | LGGTETEYSLLHMPS | |
| GluR2 | P42262 | Y876 | YKEGYNVYGIESVKI | |
| GRP58 | P30101 | Y445 | ANDVPSPYEVRGFPT | |
| GRP58 | P30101 | Y454 | VRGFPTIYFSPANKK | |
| GRP58 | P30101 | Y467 | KKLNPKKYEGGRELS | |
| HS1 (HCLS1) | P14317 | Y222 | MEAPTTAYKKTTPIE | + |
| LAB | Q9GZY6 | Y110 | RHGSEEAYIDPIAME | |
| LAB | Q9GZY6 | Y118 | IDPIAMEYYNWGRFS | |
| LAB | Q9GZY6 | Y119 | DPIAMEYYNWGRFSK | |
| LAB | Q9GZY6 | Y136 | EDDDANSYENVLICK | |
| LAB | Q9GZY6 | Y95 | EDPASSRYQNFSKGS | |
| LPXN | O60711 | Y72 | NIQELNVYSEAQEPK | |
| Lyn | P07948 | Y397 | RVIEDNEYTAREGAK | + |
| NIPP-1 | Q12972 | Y335 | NEPKKKKYAKEAWPG | |
| NMT1 | P30419 | Y117 | EEASKRSYQFWDTQP | |
| NMT1 | P30419 | Y180 | YTLLNENYVEDDDNM | + |
| p27Kip1 | P46527 | T187 | NAGSVEQTPKKPGLR | + |
| p27Kip1 | P46527 | Y88 | KGSLPEFYYRPPRPP | ? |
| PAG (PAG1) | Q9NWQ8 | Y381 | GEEPEPDYEAIQTLN | |
| PAG (PAG1) | Q9NWQ8 | Y387 | SEEPEPDYEAIQTLN | |
| PAG (PAG1) | Q9NWQ8 | Y417 | LVPKENDYESISDLQ | |
| PAG (PAG1) | Q9NWQ8 | Y431 | LTVPESTYTSIQGDP | |
| PKCd (PRKCD) | Q05655 | Y155 | IKQAKIHYIKNHEFI | - |
| PKCd (PRKCD) | Q05655 | Y313 | SSEPVGIYQGFEKKT | + |
| PKCd (PRKCD) | Q05655 | Y334 | MQDNSGTYGKIWEGS | ? |
| PKCd (PRKCD) | Q05655 | Y52 | VQKKPTMYPEWKSTF | - |
| PKCd (PRKCD) | Q05655 | Y567 | IRVDTPHYPRWITKE | - |
| PTEN | P60484 | Y240 | RREDKFMYFEFPQPL | + |
| PTPN6 (SHP1) | P29350 | Y564 | SKHKEDVYENLHTKN | + |
| RGS16 | O15492 | Y168 | TLMEKDSYPRFLKSP | |
| RGS16 | O15492 | Y177 | RFLKSPAYRDLAAQA | |
| SLAM/SLAMF1 | Q13291 | Y327 | ETNSITVYASVTLPE | |
| SLC4A1 | P02730 | Y21 | ENLEQEEYEDPDIPE | |
| SLC4A1 | P02730 | Y359 | AKPDSSFYKGLDLNG | |
| SLC4A1 | P02730 | Y8 | MEELQDDYEDMMEEN | |
| SLC4A1 | P02730 | Y904 | EEEGRDEYDEVAMPV | |
| SNCA | P37840 | Y125 | VDPDNEAYEMPSEEG | |
| STAT5A | P42229 | Y694 | LAKAVDGYVKPQIKQ | + |
| Syk | P43405 | Y323 | STVSFNPYEPELAPW | + |
| Syk | P43405 | Y348 | LPMDTEVYESPYADP | + |
| Syk | P43405 | Y352 | TEVYESPYADPEEIR | + |
| TIF1B | Q13263 | Y449 | PMEVQEGYGFGSGDD | + |
| TIF1B | Q13263 | Y458 | FGSGDDPYSSAEPHV | + |
| TIF1B | Q13263 | Y517 | PGSTTEDYNLIVIER | + |
| TRIM28 | Q13263 | Y458 | FGSGDDPYSSAEPHV | + |
| TRIM28 | Q13263 | Y517 | PGSTTEDYNLIVIER | + |
| TRPV4 | Q9HBA0 | Y253 | IERRCKHYVELLVAQ | |
| WASP | P42768 | Y291 | AETSKLIYDFIEDQG | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
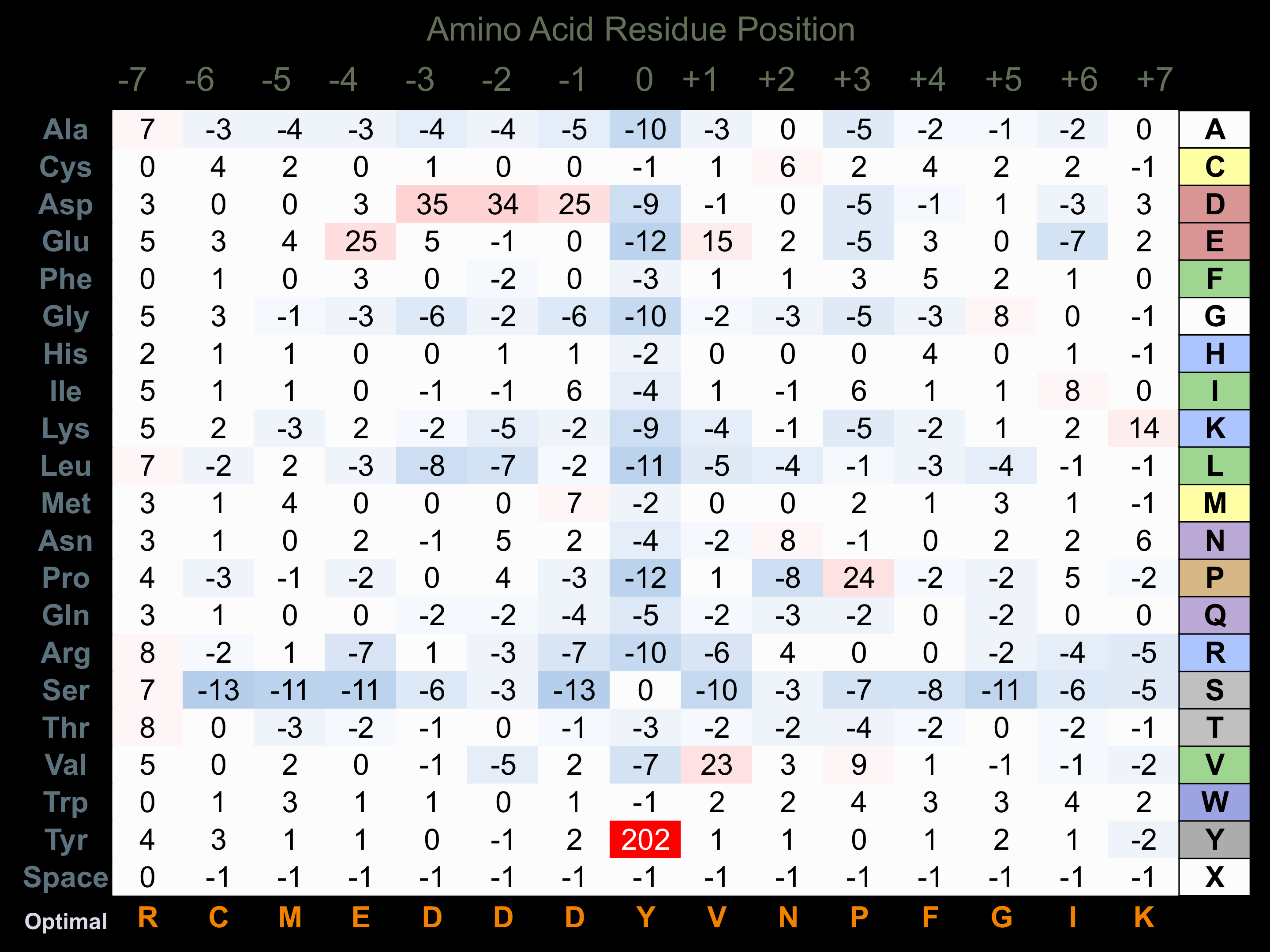
Matrix Type:
Experimentally derived from alignment of 106 known protein substrate phosphosites and 110 peptides phosphorylated by recombinant Lyn in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Chorea-Acanthocytosis; Acanthocytosis
Specific Cancer Types:
Sarcomas
Comments:
LYN appears to be an oncoprotein (OP). Lyn is constitutively phosphorylated and activated in cells of chronic myelogenous leukemia (CML) and acute myeloid leukemia (AML) patients. Abnormally enhanced expression levels of activation of Lyn signalling can be critical in survival and proliferation of some types of cancers.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +309, p<0.0001); Bladder carcinomas (%CFC= +85, p<0.002); Brain glioblastomas (%CFC= -98, p<0.002); Brain oligodendrogliomas (%CFC= -54, p<0.088); Cervical cancer (%CFC= -71, p<0.0001); Clear cell renal cell carcinomas (cRCC) (%CFC= +90, p<0.0009); Colon mucosal cell adenomas (%CFC= +47, p<0.0001); Large B-cell lymphomas (%CFC= +90, p<0.019); Oral squamous cell carcinomas (OSCC) (%CFC= +604, p<0.0001); Papillary thyroid carcinomas (PTC) (%CFC= +81, p<0.096); Pituitary adenomas (ACTH-secreting) (%CFC= -53, p<0.053); Prostate cancer - metastatic (%CFC= +46, p<0.0001); and Skin squamous cell carcinomas (%CFC= +102, p<0.0009). The COSMIC website notes an up-regulated expression score for LYN in diverse human cancers of 404, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 19 for this protein kinase in human cancers was 0.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25188 diverse cancer specimens. This rate is only 12 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.49 % in 805 skin cancers tested; 0.43 % in 1093 large intestine cancers tested; 0.32 % in 602 endometrium cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,440 cancer specimens
Comments:
Only 3 deletions, and no insertions or complex mutations are noted on the COSMIC website.