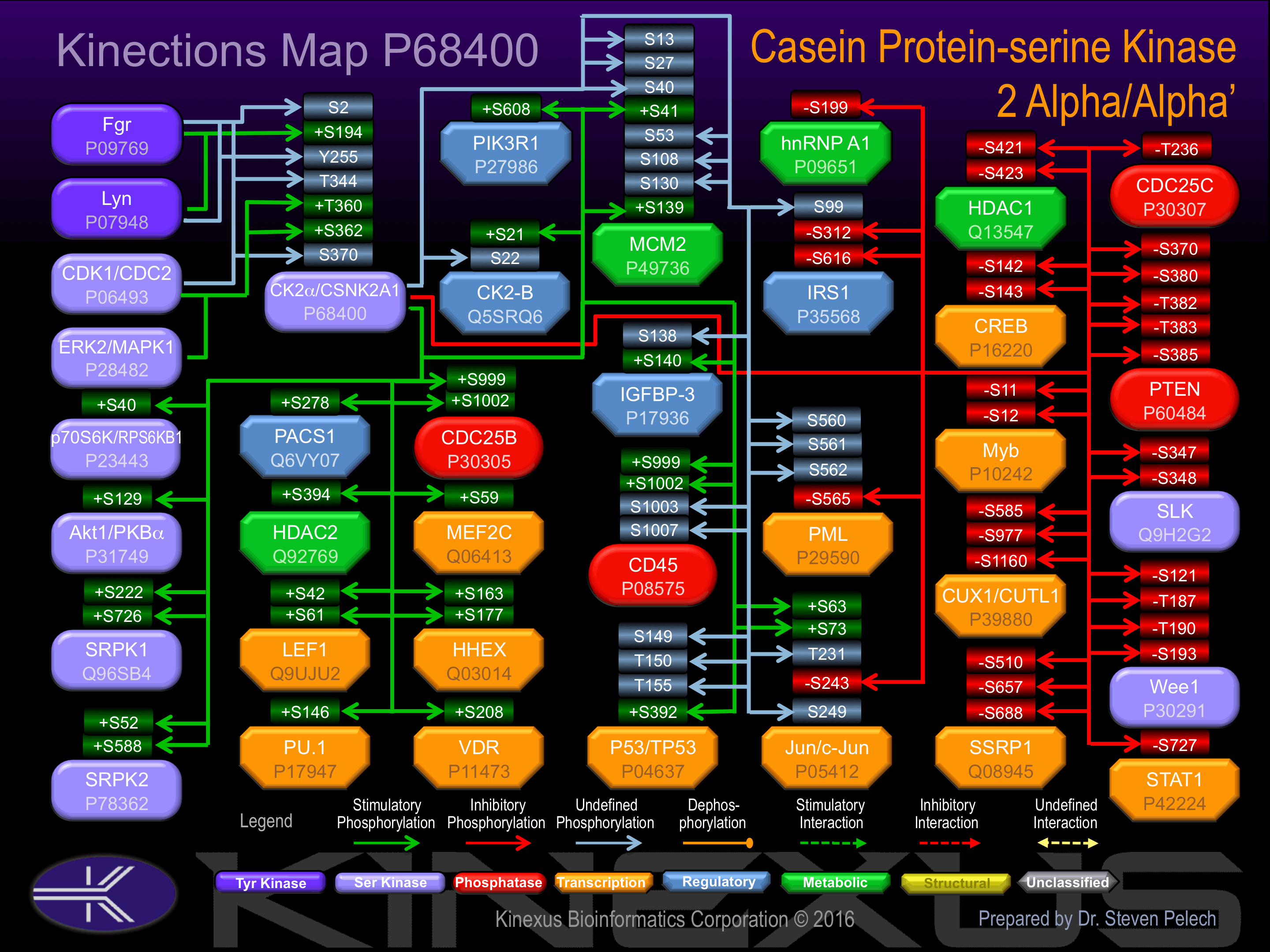
Nomenclature
Short Name:
CK2a1
Full Name:
Casein kinase II, alpha chain
Alias:
- Casein kinase 2, alpha 1 polypeptide
- Casein kinase II, alpha chain
- CSNK2A1
- EC 2.7.11.1
- Kinase CK2-alpha1
- CK II
- CK2-alpha1
- CKIIA
- CSK21
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
CK2
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
45,144
# Amino Acids:
391
# mRNA Isoforms:
2
mRNA Isoforms:
45,144 Da (391 AA; P68400); 29,181 Da (255 AA; P68400-2)
4D Structure:
Tetramer composed of an alpha chain, an alpha' and two beta chains. Also component of a CK2-SPT16-SSRP1 complex composed of SSRP1, SUPT16H, CSNK2A1, CSNK2A2 and CSNK2B, the complex associating following UV irradiation. Interacts with RNPS1.
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 39 | 324 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K49, K102, K122, K329.
Serine phosphorylated:
S2, S7, S51, S194+, S277, S287, S362+, S370.
Threonine phosphorylated:
T13, T17, T60, T127, T344, T360+.
Tyrosine phosphorylated:
Y12, Y50, Y182, Y239, Y255.
Ubiquitinated:
K49, K102, K122, K229, K247, K260, K329.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 91
91
1125
63
1207
 9
9
112
22
70
 46
46
575
15
473
 41
41
515
182
792
 63
63
785
52
617
 8
8
100
129
63
 22
22
275
67
474
 100
100
1243
57
2719
 54
54
676
24
513
 11
11
131
161
105
 15
15
192
43
226
 58
58
718
279
671
 19
19
230
48
319
 7
7
91
18
44
 22
22
278
32
307
 7
7
90
33
67
 19
19
237
226
1404
 28
28
345
27
333
 20
20
244
165
330
 51
51
638
218
632
 28
28
342
32
380
 22
22
271
37
330
 33
33
416
26
422
 43
43
535
28
528
 28
28
344
32
403
 92
92
1140
115
1942
 20
20
254
51
287
 25
25
307
28
330
 25
25
311
28
322
 13
13
162
70
116
 58
58
723
42
577
 86
86
1065
67
2288
 29
29
364
114
674
 70
70
874
135
709
 16
16
202
83
313
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
91 90.4
90.4
92.1
100 -
-
-
100 -
-
-
99 97
97
97
100 -
-
-
- 97.4
97.4
98.7
99 98.4
98.4
98.9
98.5 -
-
-
- 75.4
75.4
82.6
- 97.9
97.9
98.9
98 94.9
94.9
97.1
94 89
89
93.8
81 -
-
-
- 75.9
75.9
81.8
89 79.5
79.5
86.1
- 72.8
72.8
82.3
79 78.3
78.3
87.1
- -
-
-
- 65.2
65.2
74.9
- -
-
-
76 63.4
63.4
75.4
75.5 49.8
49.8
63.6
60 61.1
61.1
72.8
71
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CHEK1 - O14757 |
| 2 | HCLS1 - P14317 |
| 3 | SPIB - Q01892 |
| 4 | XRCC1 - P18887 |
| 5 | PACS1 - Q6VY07 |
| 6 | SSRP1 - Q08945 |
| 7 | HSP90B1 - P14625 |
| 8 | CFDP1 - Q9UEE9 |
| 9 | HDAC3 - O15379 |
| 10 | NFKBIA - P25963 |
| 11 | MYCN - P04198 |
| 12 | AIP - O00170 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| LYN | P07948 | S2 | ______MSGPVPSRA | |
| FGR | P09769 | S2 | ______MSGPVPSRA | |
| CK2a1 | P68400 | Y182 | DWGLAEFYHPGQEYN | |
| LYN | P07948 | S194 | EYNVRVASRYFKGPE | + |
| FGR | P09769 | S194 | EYNVRVASRYFKGPE | + |
| LYN | P07948 | Y255 | VLGTEDLYDYIDKYN | |
| FGR | P09769 | Y255 | VLGTEDLYDYIDKYN | |
| CDK1 | P06493 | T344 | SSMPGGSTPVSSANM | |
| ERK2 | P28482 | T360 | SGISSVPTPSPLGPL | + |
| CDK1 | P06493 | T360 | SGISSVPTPSPLGPL | + |
| ERK2 | P28482 | S362 | ISSVPTPSPLGPLAG | + |
| CDK1 | P06493 | S362 | ISSVPTPSPLGPLAG | + |
| CDK1 | P06493 | S370 | PLGPLAGSPVIAAAN |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 4E-BP1 | Q13541 | S111 | KRAGGEESQFEMDI_ | |
| 4E-BP1 | Q13541 | S64 | FLMECRNSPVTKTPP | |
| A-Myb | P10243 | S7 | _MAKRSRSEDEDDDL | |
| ABCA1 | O95477 | S1255 | FLKVAEESGVDAETS | |
| ABCA1 | O95477 | T1242 | SSYGISETTLEEIFL | |
| ABCA1 | O95477 | T1243 | SYGISETTLEEIFLK | |
| ABCF1 | Q8NE71 | T108 | LKKLSVPTSDEEDEV | |
| ACC1 | Q13085 | S29 | GSVSEDNSEDEISNL | |
| ACE | P12821 | S1299 | SHGPQFGSEVELRHS | |
| ADNP | Q9H2P0 | S953 | TKLMHNASDSEVDQD | |
| ADNP | Q9H2P0 | S955 | LMHNASDSEVDQDDV | |
| Adrenodoxin | P10109 | T131 | YEKLDAITDEENDML | |
| Akt1 (PKBa) | P31749 | S129 | SGSPSDNSGAEEMEV | + |
| Amphiphysin | P49418 | T350 | PEVKKEETLLDLDFD | |
| Amphiphysin | P49418 | T387 | LPWDLWTTSTDLVQP | |
| ANP32A | P39687 | S158 | DDKEAPDSDAEGYVE | |
| ANP32A | P39687 | S17 | ELRNRTPSDVKELVL | |
| ANP32A | P39687 | S204 | EGEEEDVSGEEEEDE | |
| ANP32A | P39687 | S77 | ELSDNRVSGGLEVLA | |
| ANP32A | P39687 | T15 | HLELRNRTPSDVKEL | |
| ANP32A | P39687 | Y148 | QLTYLDGYDRDDKEA | |
| ANP32B | Q92688 | T244 | GEKRKRETDDEGEDD | |
| ApoE | P02649 | S314 | TSAAPVPSDNH____ | |
| AQP4 | P55087 | S276 | AAQQTKGSYMEVEDN | |
| AQP4 | P55087 | S285 | MEVEDNRSQVETDDL | |
| ARC | O60936-2 | T149 | SEAVQSGTPEEPEPE | |
| ARNT | P27540 | S77 | DKERFARSDDEQSSA | |
| ARRB2 | P32121 | T382 | EFDTNYATDDDIVFE | |
| ASCL1 | P50553 | S155 | GAANKKMSKVETLRS | |
| ATF1 | P18846 | S36 | AQQVSSLSESEESQD | |
| ATG4B | Q9Y4P1 | S383 | RLERFFDSEDEDFEI | |
| Bad | Q92934 | T80 | HSSYPAGTEDDEGMG | |
| Bamacan | Q9UQE7 | S1067 | GDVEGSQSQDEGEGS | |
| BCAM (Lu) | P50895 | S598 | GEPGLSHSGSEQPEQ | |
| BCLAF1 | Q9NYF8 | S385 | QEALDYFSDKESGKQ | |
| Bid | P55957 | T59 | EGYDELQTDGNRSSH | |
| Bik | Q13323 | S35 | EVLGMTDSEEDLDPM | |
| Bik | Q13323 | T33 | TMEVLGMTDSEEDLD | |
| BMAL1 | O00327 | S90 | RRRDKMNSFIDELAS | |
| BRCA1 | P38398 | S1572 | ESGISLFSDDPESDP | |
| BYSL | Q13895 | S98 | PRMPQDGSDDEDEEW | |
| C1R | P00736 | S206 | TEASGYISSLEYPRS | |
| CADPS | Q9ULU8 | S1345 | QGISMKDSDEEDEED | |
| CADPS | Q9ULU8 | S5 | MLDPSSSEEESD___ | |
| CADPS | Q9ULU8 | S6 | __MLDPSSSEEESDE | |
| CADPS | Q9ULU8 | S7 | MLDPSSSEEESDEI_ | |
| Caldesmon | Q05682 | S73 | VEVNAQNSVPDEEAK | |
| Calmodulin | P62158 | S101 | KDGNGYISAAELRHV | |
| Calmodulin | P62158 | S81 | RKMKDTDSEEEIREA | |
| Calmodulin | P62158 | T117 | TNLGEKLTDEEVDEM | |
| Calmodulin | P62158 | T79 | MARKMKDTDSEEEIR | |
| Calsequestrin 1 | P31415 | T381 | VLEGEINTEDDDDDD | |
| Calsequestrin 2 | O14958 | S385 | DDDDDDNSDEEDNDD | |
| Calsequestrin 2 | O14958 | S393 | DEEDNDDSDDDDDE_ | |
| CAPZA1 | P52907 | S9 | ADFDDRVSDEEKVRI | |
| CARD9 | Q9H257 | T531 | NTTGSDNTDTEGS__ | |
| CARD9 | Q9H257 | T533 | TGSDNTDTEGS____ | |
| Caspase 2 | P42575 | S140 | LYKKLRLSTDTVEHS | |
| Caspase 9 | P55211 | S310 | PEDESPGSNPEPDAT | |
| Caveolin 1 (CAV1) | Q03135 | S88 | FDGIWKASFTTFTVT | |
| Caveolin 2 (CAV2) | P51636 | S36 | DPEKFADSDQDRDPH | |
| CBX | P83916 | T51 | GFSDEDNTWEPEENL | |
| CBX3 | Q13185 | S95 | GTKRKSLSDSESDDS | |
| CCDC132 | Q96JG6 | S561 | YQEYDSDSDVPEELK | |
| CD45 | P08575 | S1002 | SEHDSDESSDDDSDS | + |
| CD45 | P08575 | S1003 | EHDSDESSDDDSDSE | |
| CD45 | P08575 | S1007 | DESSDDDSDSEEPSK | |
| CD45 | P08575 | S999 | SKESEHDSDESSDDD | + |
| CD5 | P06127 | S482 | SSMQPDNSSDSDYDL | |
| CD5 | P06127 | S483 | SMQPDNSSDSDYDLH | |
| CD5 | P06127 | S485 | QPDNSSDSDYDLHGA | |
| Cdc25B | P30305 | S186 | EAGSGAASSSGEDKE | + |
| Cdc25B | P30305 | S187 | AGSGAASSSGEDKEN | + |
| Cdc25C | P30307 | T236 | VEKFKDNTIPDKVKK | - |
| Cdc34 | P49427 | S203 | APAPDEGSDLFYDDY | |
| Cdc34 | P49427 | S222 | EVEEEADSCFGDDED | |
| Cdc34 | P49427 | S231 | FGDDEDDSGTEES__ | |
| Cdc34 | P49427 | S236 | DDSGTEES_______ | |
| Cdc34 | P49427 | T233 | DDEDDSGTEES____ | |
| Cdc37 | Q16543 | S13 | VWDHIEVSDDEDETH | |
| CDH1 | P12830 | S838 | LVFDYEGSGSEAASL | |
| CDH1 | P12830 | S847 | SEAASLSSLNSSESD | |
| CDH1 | P12830 | S850 | ASLSSLNSSESDKDQ | |
| CDH1 | P12830 | S851 | SLSSLNSSESDKDQD | |
| CDH1 | P12830 | S853 | SSLNSSESDKDQDYD | |
| CDK1 (CDC2) | P06493 | S39 | MKKIRLESEEEGVPS | ? |
| CEP170 | Q5SW79 | S1112 | RARLGEASDSELADA | |
| CFTR | P13569 | S422 | NNNNRKTSNGDDSLF | |
| CFTR | P13569 | S511 | ENIIFGVSYDEYRYR | |
| CFTR | P13569 | S670 | TETLHRFSLEGDAPV | |
| CHMP7 | Q8WUX9 | S417 | SVPNPRISDAELEAE | |
| CHOP | P35638 | S14 | PFSFGTLSSWELEAW | |
| CHOP | P35638 | S15 | FSFGTLSSWELEAWY | |
| CHOP | P35638 | S30 | EDLQEVLSSDENGGT | |
| CHOP | P35638 | S31 | DLQEVLSSDENGGTY | |
| CK2-B | Q5SRQ6 | S21 | TSADVKMSSSEEVSW | + |
| CK2-B | Q5SRQ6 | S22 | SADVKMSSSEEVSWI | |
| CK2a1 (CSNK2A1) | P68400 | Y182 | DWGLAEFYHPGQEYN | |
| CLTB | P09497 | S11 | DFGFFSSSESGAPEA | |
| CLTB | P09497 | S13 | GFFSSSESGAPEAAE | |
| CPLX1 | O14810 | S115 | EVEEEDESILDTVIK | |
| CPLX2 | Q6PUV4 | S115 | EEEEEEESILDTVLK | |
| CPSF2 | Q9P2I0 | S419 | SKEADIDSSDESDIE | |
| CPSF2 | Q9P2I0 | S420 | KEADIDSSDESDIEE | |
| CPT1A | P50416 | S741 | FHISSKFSCPETDSH | |
| CPT1A | P50416 | S747 | FSCPETDSHRFGRHL | |
| CREB1 | P16220 | S108 | QISTIAESEDSQESV | ? |
| CREB1 | P16220 | S111 | TIAESEDSQESVDSV | - |
| CREB1 | P16220 | S114 | ESEDSQESVDSVTDS | ? |
| CREB1 | P16220 | S117 | DSQESVDSVTDSQKR | |
| CREB1 | P16220 | S121 | SVDSVTDSQKRREIL | |
| CREB1 | P16220 | S142 | RKILNDLSSDAPGVP | - |
| CREB1 | P16220 | S143 | KILNDLSSDAPGVPR | - |
| CREM | Q03060-2 | S94 | PKIEEERSEEEGTPP | |
| CREM | Q03060-2 | T47 | QVAAIAETDESAESE | |
| CSIG | O76021 | S361 | QVKATNESEDEIPQL | |
| CstF-77 | Q12996 | S691 | VKRPNEDSDEDEEKG | |
| CTCF | P49711 | S604 | GRKRKMRSKKEDSSD | |
| CTCF | P49711 | S609 | MRSKKEDSSDSENAE | |
| CTCF | P49711 | S610 | RSKKEDSSDSENAEP | |
| CTCF | P49711 | S612 | KKEDSSDSENAEPDL | |
| CTNNB1 | P35222 | T393 | RNLSDAATKQEGMEG | |
| CUX1 (CUTL1) | P39880 | S1160 | ILGLTQGSVSDLLAR | - |
| CUX1 (CUTL1) | P39880 | S585 | VLGLSQGSVSEILAR | - |
| CUX1 (CUTL1) | P39880 | S977 | VLGLSQGSVSDMLSR | - |
| Cx50 | P48165 | S387 | GEKEEPQSEKVSKQG | |
| Cyclin H (CCNH) | P51946 | T315 | KHEEEEWTDDDLVES | |
| Cyclin H (CCNH) | P51946 | Y99 | LNNSVMEYHPRIIML | |
| DARPP-32 | Q9UD71 | S102 | NLNENQASEEEDELG | |
| DARPP-32 | Q9UD71 | S42 | PAMLFRLSEHSSPEE | |
| DARPP-32 | Q9UD71 | S45 | LFRLSEHSSPEEEAS | |
| DDHD1 | Q8NEL9 | S104 | GSSLRYYSEGESGGG | |
| DDHD1 | Q8NEL9 | S713 | AKEPTSVSENEGIST | |
| DDHD1 | Q8NEL9 | S92 | SDENYDFSSAESGSS | |
| DDX24 | Q9GZR7 | S82 | KRKAQAVSEEEEEEE | |
| DEK | P35659 | S159 | FRNAMLKSICEVLDL | |
| DEK | P35659 | S201 | PKSKKTCSKGSKKER | |
| DEK | P35659 | S204 | KKTCSKGSKKERNSS | |
| DEK | P35659 | S243 | EKKNKEESSDDEDKE | |
| DEK | P35659 | S244 | KKNKEESSDDEDKES | |
| DEK | P35659 | S251 | SDDEDKESEEEPPKK | |
| DEK | P35659 | S287 | ANVKKADSSTTKKNQ | |
| DEK | P35659 | S288 | NVKKADSSTTKKNQN | |
| DEK | P35659 | S296 | TTKKNQNSSKKESES | |
| DEK | P35659 | S301 | QNSSKKESESEDSSD | |
| DEK | P35659 | S303 | SSKKESESEDSSDDE | |
| DEK | P35659 | S306 | KESESEDSSDDEPLI | |
| DEK | P35659 | S307 | ESESEDSSDDEPLIK | |
| DEK | P35659 | S307 | ESESEDSSDDEPLIK | |
| DEK | P35659 | S32 | MPGPREESEEEEDED | |
| DEK | P35659 | T199 | PLPKSKKTCSKGSKK | |
| DEK | P35659 | T289 | VKKADSSTTKKNQNS | |
| DEK | P35659 | T290 | KKADSSTTKKNQNSS | |
| dyskerin | O60832 | S451 | TAKRKRESESESDET | |
| eEF1B | P24534 | S105 | DDIDLFGSDDEEESE | |
| eEF1B | P24534 | S111 | GSDDEEESEEAKRLR | |
| eEF1D | P29692 | S162 | DDIDLFGSDNEEEDK | |
| eIF2B | P20042 | S2 | ______MSGDEMIFD | |
| eIF2B | P20042 | S67 | DTRKKDASDDLDDLN | |
| eIF2B-e | Q13144 | S717 | LKEAEEESSEDD___ | |
| eIF2B-e | Q13144 | S718 | KEAEEESSEDD____ | |
| eIF3C | Q99613 | S39 | GKQPLLLSEDEEDTK | |
| eIF5 | P55010 | S174 | DKENGSVSSSETPPP | |
| eIF5 | P55010 | S389 | LKEAEEESSGGEEED | |
| eIF5 | P55010 | S390 | KEAEEESSGGEEEDE | |
| eIF5 | P55010 | T207 | DDDWGEDTTEEAQRR | |
| eIF5 | P55010 | T208 | DDWGEDTTEEAQRRR | |
| EIF5B | O60841 | S214 | KPGPNIESGNEDDDA | |
| ERa (ESR1) | P03372 | S167 | GGRERLASTNDKGSM | ? |
| EXOSC9 | Q06265 | S392 | QDAPIILSDSEEEEM | |
| EXOSC9 | Q06265 | S394 | APIILSDSEEEEMII | |
| Factor V | P12259 | S720 | LEPEDEESDADYDYQ | |
| FAF1 | Q9UNN5 | S289 | ITDVHMVSDSDGDDF | |
| FAF1 | Q9UNN5 | S291 | DVHMVSDSDGDDFED | |
| FAM133B | Q5BKY9 | S196 | PLSSESLSESEYIEE | |
| FETUA | P02765 | S325 | HTFMGVVSLGSPSGE | |
| FETUA | P02765 | S328 | MGVVSLGSPSGEVSH | |
| FGA | P02671 | S542 | PMLGEFVSETESRGS | |
| FGA | P02671 | S609 | KMADEAGSEADHEGT | |
| FKBP15 | Q5T1M5 | S1164 | HSQRSSLSGDEEDEL | |
| FKBP4 | Q02790 | T143 | EFKGEDLTEEEDGGI | |
| FMR1 | Q06787 | S500 | NSEASNASETESDHR | |
| FNBP3 | O75400 | S938 | DTSGSELSEGELEKR | |
| FosB | P53539 | S27 | SAESQYLSSVDSFGS | |
| G3BP-1 | Q13283 | S149 | VTEPQEESEEEVEEP | |
| G6PI | P06744 | S184 | GPRVWYVSNIDGTHI | |
| GADD45GIP1 | Q8TAE8 | S221 | PAASGAPSS______ | |
| GAIP | P49795 | S24 | ADRPPSMSSHDTASP | |
| GAP43 | P17677 | S192 | PTETAESSQAEEEKE | |
| GAP43 | P17677 | S202 | PPTETGESSQAEENI | |
| GAP43 | P17677 | S203 | PTETGESSQAEENIE | |
| GAP43 | P17677 | T86 | EKKGEGTTTAEAAPA | |
| GAP43 | P17677 | T87 | KKGEGTTTAEAAPAT | |
| GAP43 | P17677 | T94 | TAEAAPATGSKPDEP | |
| GBF1 | Q92538 | S1318 | SLDRGYTSDSEVYTD | |
| GGA1 | Q9UJY5 | S355 | EQPSASVSLLDDELM | |
| GMFb | P60983 | S52 | DEELEGISPDELKDE | |
| GYS1 | P13807 | S656 | RHSSPHQSEDEEEPR | |
| HDAC1 | Q13547 | S393 | EDAIPEESGDEDEDD | |
| HDAC1 | Q13547 | S421 | IACEEEFSDSEEEGE | - |
| HDAC1 | Q13547 | S423 | CEEEFSDSEEEGEGG | - |
| HDAC2 | Q92769 | S394 | EDAVHEDSGDEDGED | + |
| HDAC2 | Q92769 | S424 | CDEEFSDSEDEGEGG | |
| HDAC3 | O15379 | S424 | DHDNDKESDVEI___ | |
| HDGF | P51858 | S132 | KKGNAEGSSDEEGKL | |
| HES6 | Q96HZ4 | S183 | GPGDDLCSDLEEAPE | |
| HHEX | Q03014 | S163 | FETQKYLSPPERKRL | + |
| HHEX | Q03014 | S177 | LAKMLQLSERQVKTW | + |
| HIRIP3 | Q9BW71 | S142 | ASKAVEESSDEERQR | |
| HIRIP3 | Q9BW71 | S143 | SKAVEESSDEERQRD | |
| HMGA1 | P17096 | S102 | EEGISQESSEEEQ__ | |
| HMGA1 | P17096 | S103 | EGISQESSEEEQ___ | |
| HMGN1 (HMG14) | P05114 | S6 | __PKRKVSSAEGAAK | |
| HMGN1 (HMG14) | P05114 | S7 | _PKRKVSSAEGAAKE | |
| HMGN1 (HMG14) | P05114 | S85 | GETKTEESPASDEAG | |
| HMGN1 (HMG14) | P05114 | S88 | KTEESPASDEAGEKE | |
| HMGN1 (HMG14) | P05114 | S98 | AGEKEAKSD______ | |
| hnRNP A1 | P09651 | S199 | SQRGRSGSGNFGGGR | - |
| hnRNP C1,C2 | P07910 | S260 | SEGGADDSAEEGDLL | |
| Hox-B6 | P17509 | S214 | LLSASQLSAEEEEEK | |
| Hox-B7 | P09629 | S132 | RIYPWMRSSGTDRKR | ? |
| Hox-B7 | P09629 | S133 | IYPWMRSSGTDRKRG | ? |
| Hox-B7 | P09629 | T203 | NKTAGPGTTGQDRAE | ? |
| Hox-B7 | P09629 | T204 | KTAGPGTTGQDRAEA | ? |
| HP1 alpha | P45973 | S11 | KTKRTADSSSSEDEE | |
| HP1 alpha | P45973 | S12 | TKRTADSSSSEDEEE | |
| HP1 alpha | P45973 | S13 | KRTADSSSSEDEEEY | |
| HS1 (HCLS1) | P14317 | T16 | DVSVSVETQGDDWDT | |
| HS1 (HCLS1) | P14317 | T23 | TQGDDWDTDPDFVND | |
| HSP105 | Q92598 | S509 | PTEENEMSSEADMEC | |
| HSP90AA1 | P07900 | S263 | PEIEDVGSDEEEEKK | |
| HSP90B | P08238 | S226 | KEREKEISDDEAEEE | ? |
| HSP90B | P08238 | S255 | PKIEDVGSDEEDDSG | ? |
| HSPC148 | Q9P013 | T110 | LDADDPLTDEEDEDF | |
| ICLN | P54105 | S102 | ADEEEEDSDDDVEPI | |
| IFI-16 | Q16666 | S132 | GAQKRKKSTKEKAGP | |
| IGF2R | P11717 | S2409 | LHGDDQDSEDEVLTI | |
| IGF2R | P11717 | S2484 | LVSFHDDSDEDLLHI | |
| IGFBP-3 | P17936 | S138 | PPAPGNASESEEDRS | |
| IGFBP-3 | P17936 | S140 | APGNASESEEDRSAG | + |
| Ikaros | Q13422 | S101 | GSHRDQGSSALSGVG | |
| Ikaros | Q13422 | S13 | GQDMSQVSGKESPPV | |
| Ikaros | Q13422 | S295 | LSDTPYDSSASYEKE | |
| Ikaros | Q13422 | S393 | REASPSNSCQDSTDT | |
| Ikaros | Q13422 | S63 | NVKVETQSDEENGRA | |
| Ikaros | Q13422 | T23 | ESPPVSDTPDEGDEP | |
| Ikaros | Q13422 | T398 | SNSCQDSTDTESNNE | |
| IkBa | P25963 | S283 | NLQMLPESEDEESYD | |
| IkBa | P25963 | S288 | PESEDEESYDTESEF | |
| IkBa | P25963 | S293 | EESYDTESEFTEFTE | - |
| IkBa | P25963 | S32 | LLDDRHDSGLDSMKD | - |
| IkBa | P25963 | S36 | RHDSGLDSMKDEEYE | - |
| IkBa | P25963 | T291 | EDEESYDTESEFTEF | - |
| IkBa | P25963 | T299 | ESEFTEFTEDELPYD | - |
| IRS1 | P35568 | S312 | TESITATSPASMVGG | - |
| IRS1 | P35568 | S616 | DDGYMPMSPGVAPVP | - |
| IRS1 | P35568 | S99 | HFAIAADSEAEQDSW | |
| ITCH | Q96J02 | T464 | VNHNTRITQWEDPRS | |
| IWS1 | Q96ST2 | S196 | EPPRHQASDSENEEP | |
| IWS1 | Q96ST2 | S400 | KAAVLSDSEDEEKAS | |
| IWS1 | Q96ST2 | S438 | KREKTIASDSEEEAG | |
| JNK2 (MAPK9) | P45984 | S407 | STEQTLASDTDSSLD | |
| JNK2 (MAPK9) | P45984 | T404 | SSMSTEQTLASDTDS | |
| Jun (c-Jun) | P05412 | S243 | PGETPPLSPIDMESQ | - |
| Jun (c-Jun) | P05412 | S249 | LSPIDMESQERIKAE | |
| Jun (c-Jun) | P05412 | S63 | KNSDLLTSPDVGLLK | + |
| Jun (c-Jun) | P05412 | S73 | VGLLKLASPELERLI | + |
| Jun (c-Jun) | P05412 | T231 | ALKEEPQTVPEMPGE | |
| KANSL2 | Q9H9L4 | S175 | SEADSIDSDQEDPLK | |
| KIR3DL1 | P43629 | S385 | AGNRTANSEDSDEQD | + |
| KIR3DL1 | P43629 | S388 | RTANSEDSDEQDPEE | + |
| KLF1 | Q13351 | T23 | ALGPFPDTQDDFLKW | |
| KPNA3 | O00505 | S60 | QEESLEDSDVDADFK | |
| KRI1 | Q8N9T8 | S177 | FRAFVEDSEDEDGAG | |
| KRI1 | Q8N9T8 | S287 | QLAVDDSSDEGELFL | |
| LDLR | P01130 | S854 | YPSRQMVSLEDDVA_ | |
| LEDGF | O75475 | S106 | TKQSNASSDVEVEEK | |
| LEDGF | O75475 | S275 | GVTSTSDSEEEGDDQ | |
| LEF1 | Q9UJU2 | S42 | EKIFAEISHPEEEGD | + |
| LEF1 | Q9UJU2 | S61 | KSSLVNESEIIPASN | + |
| LEO1 | Q8WVC0 | S630 | LKAKKLTSDEEGEPS | |
| LIG1 | P18858 | S66 | KAARVLGSEGEEEDE | |
| M6PR | P20645 | S267 | DDQLGEESEERDDHL | |
| MAP1A | P78559 | S2106 | SHWDDSTSDSELEKG | |
| MAX | P61244 | S11 | NDDIEVESDEEQPRF | |
| MAX | P61244 | S2 | ______MSDNDDIEV | |
| MAZ | P56270 | S460 | PTAVGSLSGAEGVPV | |
| MCM2 | P49736 | S108 | DVEELTASQREAAER | + |
| MCM2 | P49736 | S13 | ESFTMASSPAQRRRG | ? |
| MCM2 | P49736 | S130 | RRGLLYDSDEEDEER | |
| MCM2 | P49736 | S139 | RRGLLYDSDEEDEER | + |
| MCM2 | P49736 | S27 | GNDPLTSSPGRSSRR | ? |
| MCM2 | P49736 | S40 | RRTDALTSSPGRDLP | ? |
| MCM2 | P49736 | S41 | RTDALTSSPGRDLPP | + |
| MCM2 | P49736 | S53 | LPPFEDESEGLLGTE | ? |
| MCM3 | P25205 | S672 | KKRSEDESETEDEEE | |
| MCM3 | P25205 | S711 | SYDPYDFSDTEEEMP | |
| MDC1 | Q14676 | S218 | PFAFNLNSDTDVEEG | |
| MDC1 | Q14676 | S299 | SQPPGEDSDTDVDDD | |
| MDC1 | Q14676 | S329 | QPFGFIDSDTDAEEE | |
| MDC1 | Q14676 | S376 | LQESQAGSDTDVEEG | |
| MDC1 | Q14676 | S402 | QASMVINSDTDDEEE | |
| MDC1 | Q14676 | S453 | QTTTERDSDTDVEEE | |
| MDC1 | Q14676 | T220 | AFNLNSDTDVEEGQQ | |
| MDC1 | Q14676 | T301 | PPGEDSDTDVDDDSR | |
| MDC1 | Q14676 | T331 | FGFIDSDTDAEEERI | |
| MDC1 | Q14676 | T378 | ESQAGSDTDVEEGKA | |
| MDC1 | Q14676 | T404 | SMVINSDTDDEEEVS | |
| MDC1 | Q14676 | T455 | TTERDSDTDVEEEEL | |
| MDM2 | Q00987 | S260 | SLDSEDYSLSEEGQE | |
| MDM2 | Q00987 | S269 | SEEGQELSDEDDEVY | |
| MEF2A | Q02078 | S289 | KGMMPPLSEEEELEL | |
| MEF2C | Q06413 | S59 | NKLFQYASTDMDKVL | + |
| MFAP1 | P55081 | S118 | EVVGESDSEVEGDAW | |
| MFAP1 | P55081 | S52 | PDYAPMESSDEEDEE | |
| MFAP1 | P55081 | S53 | DYAPMESSDEEDEEF | |
| MFAP1 | P55081 | T267 | AALDALNTDDENDEE | |
| MRE11A | P49959 | S649 | EVIEVDESDVEEDIF | |
| MRLC2 (MYL12B) | P19105 | T135 | TTMGDRFTDEEVDEL | |
| Myb | P10242 | S11 | RPRHSIYSSDEDDED | - |
| Myb | P10242 | S12 | PRHSIYSSDEDDEDF | - |
| Myc | P01106 | S249 | EETPPTTSSDSEEEQ | |
| Myc | P01106 | S250 | ETPPTTSSDSEEEQE | |
| Myc | P01106 | S252 | PPTTSSDSEEEQEDE | |
| Myc | P01106 | S347 | RKCTSPRSSDTEENV | |
| Myc | P01106 | S348 | KCTSPRSSDTEENVK | |
| Myf-5 | P13349 | S133 | NAIRYIESLQELLRE | |
| Myf-5 | P13349 | S49 | HKAELQGSDEDEHVR | |
| MYH10 | P35580 | S1952 | QLHLEGASLELSDDD | |
| MYH10 | P35580 | S1956 | EGASLELSDDDTESK | |
| MYH10 | P35580 | S1965 | DDTESKTSDVNETQP | |
| MYH10 | P35580 | S1975 | NETQPPQSE______ | |
| MYH10 | P35580 | T1960 | LELSDDDTESKTSDV | |
| MYH13 | Q9UKX3 | S2 | ______MSSDAEMAI | |
| MYH13 | Q9UKX3 | S3 | _____MSSDAEMAIF | |
| MYH2 | Q9UKX2 | S1921 | ERADIAESQVNKLRV | |
| MYH9 | P35579 | S1943 | RKGAGDGSDEEVDGK | |
| MYO18A | Q92614 | S2041 | RYSHSYLSDSDTEAK | |
| MYO18A | Q92614 | S2043 | SHSYLSDSDTEAKLT | |
| N-CAM L1 | P32004 | S1181 | GEYRSLESDNEEKAF | |
| N-Myc | P04198 | S261 | TSGEDTLSDSDDEDD | |
| N-Myc | P04198 | S263 | GEDTLSDSDDEDDEE | |
| NAP1L4 | Q99733 | S125 | DAESEWHSENEEEEK | |
| NCAPG | Q9BPX3 | S674 | CEGTEINSDDEQESK | |
| NCAPH | Q15003 | S570 | PGLQAADSDDEDLDD | |
| NCL | P19338 | S145 | DEEEDDDSEEDEEDD | |
| NCL | P19338 | S145 | KNAKKEDSDEEEDDD | |
| NCL | P19338 | S184 | AAAAAPASEDEDDED | |
| NCL | P19338 | S184 | AAAAAPASEDEDDED | |
| NCL | P19338 | S206 | DDDEEDDSEEEAMET | |
| NCL | P19338 | S206 | DDDEEDDSEEEAMET | |
| NCL | P19338 | S34 | DSEDEEMSEDEEDDS | |
| NCOR2 | Q9Y618 | S2522 | CSQYETLSDSE____ | |
| NFAT2 | O95644 | S187 | RSCNSEASSYESNYS | |
| NFkB-p65 | Q04206 | S529 | GLPNGLLSGDEDFSS | |
| NFL (Neurofilament L) | P07196 | S472 | EAKDEPPSEGEAEEE | |
| NHE3 | P48764 | S555 | AEGERRGSLAFIRSP | |
| NHE3 | P48764 | S718 | KEKDLELSDTEEPPN | |
| NIPP-1 | Q12972 | S204 | KNSRVTFSEDDEIIN | |
| NIPP-1 | Q12972 | T161 | LGLPEEETELDNLTE | |
| NKX2-5 | P52952 | S164 | FKQQRYLSAPERDQL | |
| NM23 (NME1) | P15531 | S120 | GRNIIHGSDSVESAE | |
| NMD3 | Q96D46 | T470 | AIPVESDTDDEGAPR | |
| NMDAR2B (GRIN2B) | Q13224 | S1482 | EKLSSIESDV | |
| NOLC1 | Q14978 | S90 | SSSDSEDSSEEEEEV | |
| NPM1 | P06748 | S125 | AVEEDAESEDEEEED | |
| NUCKS | Q9H1E3 | S19 | DYSQFQESDDADEDY | |
| NUCKS | Q9H1E3 | S58 | GKNSQEDSEDSEDKD | |
| NUCKS | Q9H1E3 | S79 | DSHSAEDSEDEKEDH | |
| NUP98 | P52948 | S888 | SKYGLQDSDEEEEEH | |
| ODC | P11926 | S303 | VLKEQTGSDDEDESS | |
| OSBP | P22059 | S351 | CSGKGDMSDEDDENE | |
| OSR1 | O95747 | S339 | EDGGWEWSDDEFDEE | |
| p27Kip1 | P46527 | S83 | WQEVEKGSLPEFYYR | |
| p47phox | P14598 | S208 | KRGWIPASFLEPLDS | |
| p47phox | P14598 | S283 | PSMYLQKSGQDVSQA | |
| p47phox | P14598 | S348 | PGPQSPGSPLEEERQ | |
| p53 (TP53) | P04637 | S149 | PVQLWVDSTPPPGTR | |
| p53 (TP53) | P04637 | S392 | FKTEGPDSD______ | + |
| p53 (TP53) | P04637 | T150 | VQLWVDSTPPPGTRV | |
| p53 (TP53) | P04637 | T155 | DSTPPPGTRVRAMAI | ? |
| p70S6K (RPS6KB1) | P23443 | S40 | DQPEDAGSEDELEEG | + |
| PACS1 | Q6VY07 | S278 | SPDIDNYSEEEEESF | + |
| PAI-RBP1 | Q8NC51 | S25 | DQLFDDESDPFEVLK | |
| PAM | P19021 | S946 | DRLSTEGSDQEKEDD | |
| PAM | P19021 | T943 | KGFDRLSTEGSDQEK | |
| PC4 | P53999 | S15 | VSSSGSDSDSEVDKK | |
| PCM-1 | Q15154 | S537 | TEESEYDSEHENSEP | |
| PDCD5 | O14737 | S119 | NRRKVMDSDEDDDY_ | |
| PDIA6 | Q15084 | S22 | LAVNGLYSSSDDVIE | |
| PDIA6 | Q15084 | S428 | VEDDIDLSDVELDDL | |
| PEA-15 | Q15121 | S116 | KDIIRQPSEEEIIKL | |
| PEDF | P36955 | S114 | ALYYDLISSPDIHGT | |
| PEDF | P36955 | S24 | SSCQNPASPPEEGSP | |
| PGRMC1 | O00264 | S181 | GEEPTVYSDEEEPKD | |
| PHF8 | Q9UPP1 | S857 | YIYPSLESDDDDPAL | |
| PhLP | Q13371 | S18 | EKLQYYYSSSEDEDS | |
| PhLP | Q13371 | S19 | KLQYYYSSSEDEDSD | |
| PhLP | Q13371 | S20 | LQYYYSSSEDEDSDH | |
| PhLP | Q13371 | S25 | SSSEDEDSDHEDKDR | |
| PhLP | Q13371 | S296 | ATCHSEDSDLEID__ | |
| PIAS1 | O75925 | S466 | VIDLTIDSSSDEEEE | |
| PIAS1 | O75925 | S467 | IDLTIDSSSDEEEEE | |
| PIAS1 | O75925 | S468 | DLTIDSSSDEEEEEP | |
| PIK3R1 | P27986 | S608 | ENTEDQYSLVEDDED | + |
| PIN4 | Q9Y237 | S19 | AGKGGAASGSDSADK | |
| PIP5K2A | P48426 | S304 | DGEEEGESDGTHPVG | |
| PKAR2A | P13861 | S77 | VADAKGDSESEEDED | |
| PKAR2A | P13861 | S79 | DAKGDSESEEDEDLE | |
| PLD1 | Q13393 | S610 | HFKLFHPSSESEQGL | |
| PLD1 | Q13393 | S611 | FKLFHPSSESEQGLT | |
| PML | P29590 | S560 | EERVVVISSSEDSDA | |
| PML | P29590 | S561 | ERVVVISSSEDSDAE | |
| PML | P29590 | S562 | RVVVISSSEDSDAEN | |
| PML | P29590 | S565 | VISSSEDSDAENSSS | - |
| POLR2F | P61218 | S2 | ______MSDNEDNFD | |
| Polycystin 2 | Q13563 | S812 | FPRSLDDSEEDDDED | |
| PPP1R2 | P41236 | S121 | YRIQEQESSGEEDSD | |
| PPP1R2 | P41236 | S122 | RIQEQESSGEEDSDL | |
| PPP1R2 | P41236 | S87 | GDDEDACSDTEATEA | |
| PPP1R7 | Q15435 | S27 | RRVESEESGDEEGKK | |
| PPPM1A | P35813 | S375 | YKNDDTDSTSTDDMW | |
| PPPM1A | P35813 | S377 | NDDTDSTSTDDMW__ | |
| PR | P06401 | S81 | TQDQQSLSDVEGAYS | ? |
| PRMT3 | O60678 | S27 | DLPELSDSGDEAAWE | |
| PRPF3 | O43395 | S619 | KGDDDEESDEEAVKK | |
| PRPF38A | Q8NAV1 | S194 | DMDDVESSEEEEEED | |
| PSEN2 | P49810 | S330 | MEEDSYDSFGEPSYP | + |
| PSEN2 | P49810 | S335 | YDSFGEPSYPEVFEP | |
| PSEN2 | P49810 | S7 | _MLTFMASDSEEEVC | |
| PSEN2 | P49810 | S9 | LTFMASDSEEEVCDE | |
| PSMA3 | P25788 | S243 | AEKYAKESLKEEDES | |
| PSMA3 | P25788 | S250 | SLKEEDESDDDNM__ | |
| PTEN | P60484 | S370 | TSVTPDVSDNEPDHY | - |
| PTEN | P60484 | S380 | EPDHYRYSDTTDSDP | - |
| PTEN | P60484 | S385 | RYSDTTDSDPENEPF | - |
| PTEN | P60484 | T382 | DHYRYSDTTDSDPEN | - |
| PTEN | P60484 | T383 | HYRYSDTTDSDPENE | - |
| PTMA | P06454 | T12 | VDTSSEITTKDLKEK | |
| PTMA | P06454 | T13 | DTSSEITTKDLKEKK | |
| PTMA | P06454 | T7 | SDAAVDTSSEITTK_ | |
| PU.1 | P17947 | S146 | QSPPLEVSDGEADGL | + |
| Rad9 | Q99638 | S341 | PKSPGPHSEEEDEAE | ? |
| Rad9 | Q99638 | S387 | SPVLAEDSEGEG___ | ? |
| RanGAP1 | P46060 | S358 | AKVLASLSDDEDEEE | |
| RBBP6 | Q7Z6E9 | S1328 | WDKDDFESEEEDVKS | |
| RBM17 | Q96I25 | S155 | ARRPDPDSDEDEDYE | |
| RBM33 | Q96EV2 | S205 | QKDIKEESDEEEEDD | |
| RBM33 | Q96EV2 | S41 | AADEDWDSELEDDLL | |
| RFC1 | P35251 | S69 | KKRIIYDSDSESEET | |
| RFC1 | P35251 | S71 | RIIYDSDSESEETLQ | |
| RFC1 | P35251 | T110 | PVTYISETDEEDDFM | |
| RNF7 | Q9UBF6 | T10 | DVEDGEETCALASHS | |
| RNPS1 | Q15287 | S53 | KGATKESSEKDRGRD | |
| RNU3IP2 | O43818 | S50 | KMNEEISSDSESESL | |
| RNU3IP2 | O43818 | S53 | NEEISSDSESESLAP | |
| RPRD2 | Q5VT52 | S374 | DVEDMELSDVEDDGS | |
| RRAD | P55042 | S214 | LVRSREVSVDEGRAC | |
| RRAD | P55042 | S257 | QIRLRRDSKEANARR | |
| RRAD | P55042 | S273 | AGTRRRESLGKKAKR | |
| RRAD | P55042 | S290 | GRIVARNSRKMAFRA | |
| RRAD | P55042 | S299 | KMAFRAKSKSCHDLS | |
| SAPLa | Q5H9R7 | S617 | QQFDDGGSDEEDIWE | |
| SAS10 | Q9NQZ2 | S368 | VTDLSDDSDFDEKAK | |
| SCN2A | Q99250 | S1112 | VPIAVGESDFENLNT | |
| SCN2A | Q99250 | S1124 | LNTEEFSSESDMEES | |
| SCN2A | Q99250 | S1126 | TEEFSSESDMEESKE | |
| SEPT2. | Q15019 | S218 | YHLPDAESDEDEDFK | |
| SF3A1 | Q15459 | S329 | EVEMEVESDEEDDKQ | |
| SF3A1 | Q15459 | S451 | RSIREKQSDDEVYAP | |
| SH2BP1 | Q6PD62 | S1021 | ISSSDDSSDEDKLKI | |
| SH2BP1 | Q6PD62 | T925 | DEFVNDDTDDDLPIS | |
| SHOX | O15266 | S106 | EKREDVKSEDEDGQT | ? |
| SIN3A | Q96ST3 | S832 | FAQRGDLSDVEEEEE | |
| SIRT1 | Q96EB6 | S659 | FHGAEVYSDSEDDVL | |
| SIRT1 | Q96EB6 | S661 | GAEVYSDSEDDVLSS | |
| SLC3A2 | P08195 | S406 | LLTSSYLSDSGSTGE | |
| SLC3A2 | P08195 | S408 | TSSYLSDSGSTGEHT | |
| SLC3A2 | P08195 | S410 | SYLSDSGSTGEHTKS | |
| SLC3A2 | P08195 | S527 | LSLFRRLSDQRSKER | |
| SLC3A2 | P08195 | S531 | RRLSDQRSKERSLLH | |
| SLK | Q9H2G2 | S347 | SSDLSIASSEEDKLS | - |
| SLK | Q9H2G2 | S348 | SDLSIASSEEDKLSQ | - |
| SLY | O75995 | S320 | LLDYDTGSEEAEEGA | |
| SNCA | P37840 | S129 | NEAYEMPSEEGYQDY | |
| SP1 | P08047 | T668 | SYCGKRFTRSDELQR | |
| SPT6 | Q7KZ85 | S73 | EGEEDEGSDSGDSED | |
| SRF | P11831 | S77 | PTAGALYSGSEGDSE | |
| SRF | P11831 | S79 | AGALYSGSEGDSESG | |
| SRF | P11831 | S83 | YSGSEGDSESGEEEE | |
| SRF | P11831 | S85 | GSEGDSESGEEEELG | |
| SRm160 | Q8IYB3 | T220 | APEPKKETESEAEDN | |
| SRPK1 | Q96SB4 | S222 | QEEEILGSDDDEQED | + |
| SRPK1 | Q96SB4 | S726 | DYLFEPHSGEEYTRD | + |
| SRPK2 | P78362 | S52 | PEEEILGSDDEEQED | + |
| SRPK2 | P78362 | S588 | DYLFEPHSGEDYSRD | + |
| SRSF11 | Q05519 | S434 | EEEQGYDSEKEKKEE | |
| SSB | P05455 | S366 | GKKTKFASDDEHDEH | |
| SSRP1 | Q08945 | S510 | SSSNEGDSDRDEKKR | - |
| SSRP1 | Q08945 | S657 | KSSSRQLSESFKSKE | - |
| SSRP1 | Q08945 | S688 | KRRRSEDSEEEELAS | - |
| Stanniocalcin-2 | O76061 | S288 | AQGPSGSSEWEDEQS | |
| STAT1 | P42224 | S727 | TDNLLPMSPEEFDEV | - |
| STLK3 | Q9UEW8 | S387 | EDGDWEWSDDEMDEK | |
| STT3B | Q8TCJ2 | S498 | ENPPVEDSSDEDDKR | |
| STT3B | Q8TCJ2 | S499 | NPPVEDSSDEDDKRN | |
| STX1A | Q16623 | S14 | ELRTAKDSDDDDDVA | |
| SYT1 | P21579 | T113 | DVKDLGKTMKDQALK | |
| SYT1 | P21579 | T129 | DDAETGLTDGEEKEE | |
| TAP | O60763 | S942 | EEEDELESGDQEDED | |
| TARDBP | Q13148 | S379 | NSYSGSNSGAAIGWG | |
| TARDBP | Q13148 | S409 | GSSMDSKSSGWGM__ | - |
| TARDBP | Q13148 | S410 | SSMDSKSSGWGM___ | - |
| Tau | P10636 | T38 | HQDQEGDTDAGLKES | |
| TCF4 | P15884 | S54 | GSNVEDRSSSGSWGN | |
| TCF4 | P15884 | S55 | SNVEDRSSSGSWGNG | |
| TCF4 | P15884 | S56 | NVEDRSSSGSWGNGG | |
| TEBP | Q15185 | S113 | WKDWEDDSDEDMSNF | |
| TEBP | Q15185 | S118 | DDSDEDMSNFDRFSE | |
| TFIP11 | Q9UBB9 | S210 | QDFPVVDSEEEAEEE | |
| TFPI | P10646 | S30 | PAPLNADSEEDEEHT | |
| TIF-IA | Q9NYV6 | S170 | KEGDVDVSDSDDEDD | |
| TIF-IA | Q9NYV6 | S172 | GDVDVSDSDDEDDNL | |
| TLE1 | Q04724 | S239 | KDSSHYDSDGDKSDD | |
| TMEM87A | Q8NBN3 | S540 | ALPALLDSDEERMIT | |
| TNNT3 | P45378 | S1 | _______SDEEVEQV | |
| TOP1 | P11387 | S10 | GDHLHNDSQIEADFR | |
| TOP2A | P11388 | S1106 | DEEENEESDNEKETE | |
| TOP2A | P11388 | S1377 | KPQKSVVSDLEADDV | |
| TOP2A | P11388 | S1469 | NRRKRKPSTSDDSDS | |
| TOP2A | P11388 | S1525 | PIKYLEESDEDDLF_ | |
| TOP2B | Q02880 | S1524 | VEAVNSDSDSEFGIP | |
| TRa | P10827 | S472 | SEADSPSSSEEEPEV | |
| TRa | P10827 | S473 | EADSPSSSEEEPEVC | |
| treacle | Q13428 | S87 | DPISTSESSEEEEEA | |
| treacle | Q13428 | S88 | PISTSESSEEEEEAE | |
| TRHR | P34981 | S383 | YLSATKVSFDDTCLA | |
| TRHR | P34981 | T365 | KESDHFSTELDDITV | |
| TRHR | P34981 | T371 | STELDDITVTDTYLS | |
| TSPY1 | Q01534 | T300 | PPEEGTETSGDSQLL | |
| TUBA3C | Q13748 | T61 | ERRPESFTTPEGPKP | |
| TUBB3 | Q13509 | S444 | YEDDEEESEAQGPK_ | |
| TXNDC1 | Q9H3N1 | S247 | EADEEDVSEEEAESK | |
| UBC3B | Q9NX64 | S233 | DCYDDDDSGNEES__ | |
| UBC3B | Q9NX64 | Y190 | VPTTLAEYCIKTKVP | |
| UFL1 | O94874 | S458 | KGRKDDDSDDESQSS | |
| VAMP4 | O75379 | S30 | RNLLEDDSDEEEDFF | |
| VDR | P11473 | S208 | SFSNLDLSEEDSDDP | + |
| VMAT2 | Q05940 | S511 | PIGEDEESESD____ | |
| VMAT2 | Q05940 | S513 | GEDEESESD______ | |
| VPS4B | O75351 | S102 | ADEKGNDSDGEGESD | |
| VTN | P04004 | T69 | VTRGDVFTMPEDEYT | |
| VTN | P04004 | T76 | TMPEDEYTVYDDGEE | |
| WASF2 | Q9Y6W5 | S482 | RRIAVEYSDSEDDSS | |
| WASF2 | Q9Y6W5 | S484 | IAVEYSDSEDDSSEF | |
| WASF2 | Q9Y6W5 | S488 | YSDSEDDSSEFDEDD | |
| WASF2 | Q9Y6W5 | S489 | SDSEDDSSEFDEDDW | |
| WASF2 | Q9Y6W5 | S497 | EFDEDDWSD______ | |
| WASP | P42768 | S483 | KRSRAIHSSDEGEDQ | |
| WASP | P42768 | S484 | RSRAIHSSDEGEDQA | |
| WDR50 | Q9Y5J1 | S121 | RVQEHEDSGDSEVEN | |
| WDR50 | Q9Y5J1 | S124 | EHEDSGDSEVENEAK | |
| Wee1 | P30291 | S121 | WEEEGFGSSSPVKSP | - |
| Wee1 | P30291 | S193 | DTPHTPKSLLSKARG | - |
| Wee1 | P30291 | T187 | RKLRLFDTPHTPKSL | - |
| Wee1 | P30291 | T190 | RLFDTPHTPKSLLSK | - |
| XPB | P19447 | S751 | FGTMSSMSGADDTVY | |
| XPG | P28715 | S563 | FDSSLLSSDDETKCK | |
| XRCC1 | P18887 | S475 | IDIEGVQSEGQDNGA | |
| XRCC1 | P18887 | S485 | QDNGAEDSGDTEDEL | |
| XRCC1 | P18887 | S518 | GEDPYAGSTDENTDS | |
| XRCC1 | P18887 | S525 | STDENTDSEEHQEPP | |
| XRCC1 | P18887 | T488 | GAEDSGDTEDELRRV | |
| XRCC1 | P18887 | T519 | EDPYAGSTDENTDSE | |
| XRCC1 | P18887 | T523 | AGSTDENTDSEEHQE | |
| XRCC4 | Q13426 | T233 | DPVYDESTDEESENQ | |
| XRN2 | Q9H0D6 | S501 | RKAEDSDSEPEPEDN | |
| ZNF265 | O95218 | S153 | KEVEDKESEGEEEDE |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
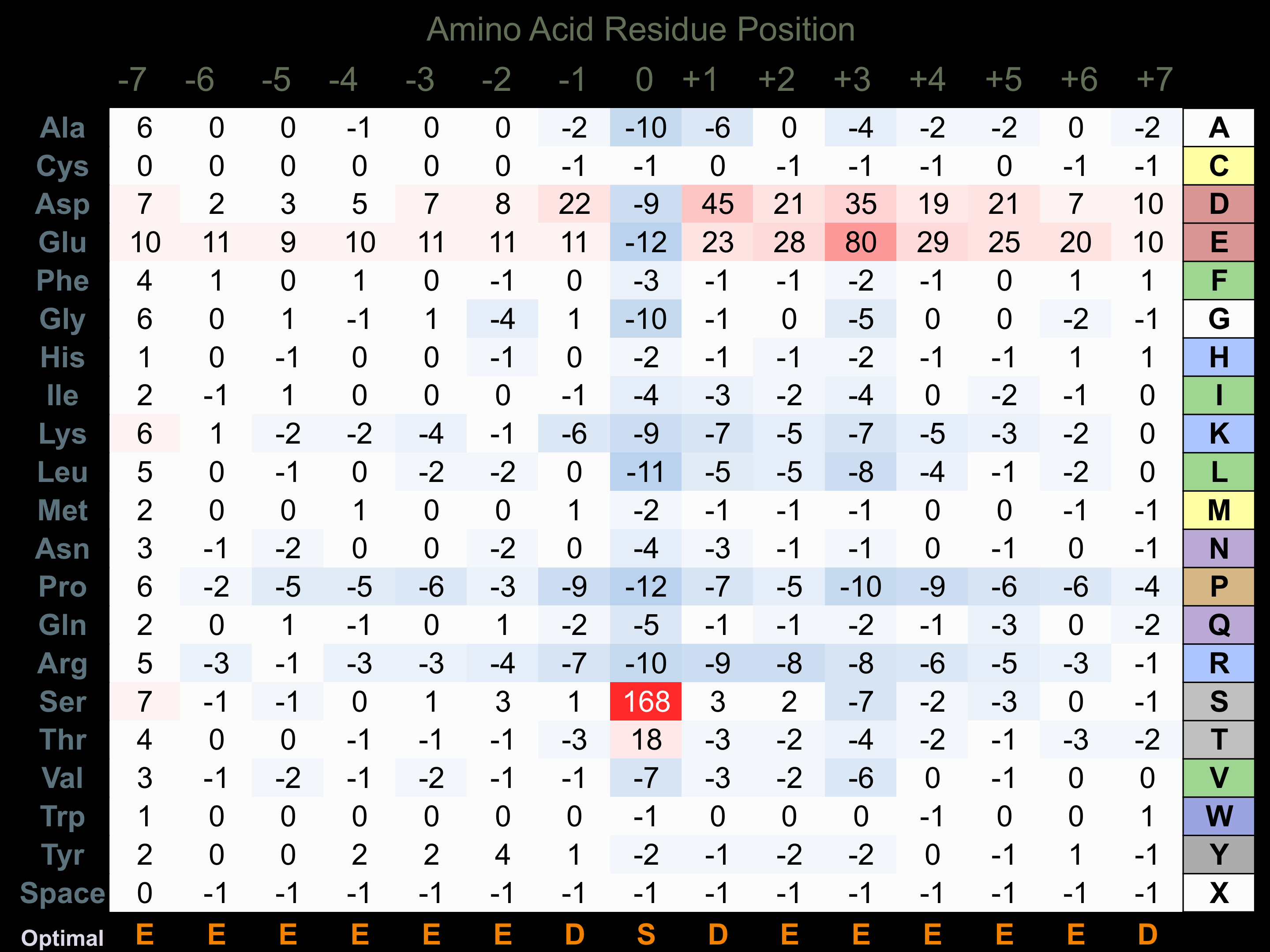
Matrix Type:
Experimentally derived from alignment of 689 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Male infertility
Specific Diseases (Non-cancerous):
Globozoospermia
Comments:
CK2a1 levels are up-regulated 2-fold in human tumours compared to most other protein kinases.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +78, p<0.0001); Brain oligodendrogliomas (%CFC= -71, p<0.009); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -91, p<0.0001); Ovary adenocarcinomas (%CFC= +72, p<0.034); Skin fibrosarcomas (%CFC= +103, p<0.014); and Skin melanomas - malignant (%CFC= +83, p<0.0006). The COSMIC website notes an up-regulated expression score for CK2a1 in diverse human cancers of 917, which is 2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 183 for this protein kinase in human cancers was 3.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25183 diverse cancer specimens. This rate is very similar (+ 6% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.3 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: RH236R (5), R80C (3). These mutations are located in the kinase catalytic domain.
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.