Nomenclature
Short Name:
PAK2
Full Name:
Serine-threonine-protein kinase PAK 2
Alias:
- C-t-PAK2
- P58
- PAK 2
- PAK-2
- S6/H4 kinase
- Gamma-PAK
- Kinase PAK2
- P21 protein (Cdc42/Rac)-activated kinase 2
- P21-activated kinase 2
- P21-activated protein kinase I
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
PAKA
Specific Links
Structure
Mol. Mass (Da):
58043
# Amino Acids:
524
# mRNA Isoforms:
1
mRNA Isoforms:
58,043 Da (524 AA; Q13177)
4D Structure:
Interacts tightly with GTP-bound but not GDP-bound CDC42/p21 and RAC1. Interacts with SH3MD4. Interacts with and activated by HIV-1 Nef. PAK-2p34 interacts with ARHGAP10. Interacts with SCRIB.
1D Structure:
Subfamily Alignment
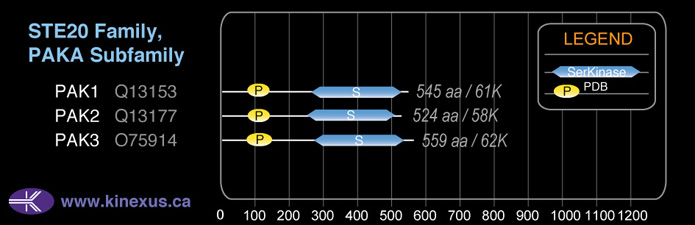
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
S2 (N-acetylserine), K28, K62 (N6), K128 (N6), K399, K492, K501.
Myristoylated:
G213 (N-myristoyl glycine in PAK2-p34 isoform).
Serine phosphorylated:
S2, S19, S20, S24, S32, S36, S42, S55, S58, S64, S75, S132, S141+, S152, S192, S197, S381, S401+, S490, S507.
Threonine phosphorylated:
T21, T25, T60, T134, T143, T154, T169, T195, T228, T394, T402+, T406.
Tyrosine phosphorylated:
Y130+, Y139, Y194, Y443, Y453.
Ubiquitinated:
K113, K136, K235, K254, K278, K299, K302, K370, K399, K468, K492, K501.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 81
81
1162
86
1060
 3
3
49
37
37
 6
6
80
6
54
 22
22
315
277
460
 57
57
820
82
549
 14
14
195
220
783
 15
15
222
109
401
 100
100
1438
83
2805
 42
42
601
38
453
 7
7
100
248
129
 5
5
76
54
80
 42
42
603
437
616
 6
6
93
61
78
 3
3
42
27
40
 5
5
68
40
73
 4
4
61
53
44
 23
23
324
554
2496
 4
4
53
23
49
 7
7
99
240
91
 42
42
598
347
557
 7
7
100
37
127
 10
10
137
46
153
 3
3
40
27
37
 4
4
54
25
40
 9
9
123
37
158
 80
80
1153
165
1910
 6
6
88
64
66
 3
3
46
25
39
 4
4
52
25
43
 7
7
101
98
131
 44
44
632
42
709
 43
43
625
96
937
 27
27
382
121
351
 65
65
937
218
734
 14
14
203
113
298
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.6
99.6
99.6
99 99
99
99.2
99 -
-
-
97 -
-
-
85 96.8
96.8
98.3
97 -
-
-
- 97.1
97.1
98.5
97 97
97
98.3
97 -
-
-
- -
-
-
- 92.8
92.8
95.6
93.5 87.2
87.2
92.6
- 83.9
83.9
89
88 -
-
-
- 34.4
34.4
50.5
- -
-
-
- 53.3
53.3
68.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 32.1
32.1
42.5
46 32.6
32.6
42.4
62
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC42 - P60953 |
| 2 | NCK1 - P16333 |
| 3 | ARHGEF7 - Q14155 |
| 4 | SRC - P12931 |
| 5 | EIF4G1 - Q04637 |
| 6 | VIM - P08670 |
| 7 | LIMK1 - P53667 |
| 8 | MYLK - Q15746 |
| 9 | MYL2 - P10916 |
| 10 | MKNK1 - Q9BUB5 |
| 11 | MAPK3 - P27361 |
| 12 | EIF4B - P23588 |
| 13 | MAPK1 - P28482 |
| 14 | RAF1 - P04049 |
| 15 | CASP3 - P42574 |
Regulation
Activation:
Activated by binding small G proteins. Binding of GTP-bound Cdc42 or Rac1 to the autoregulatory region releases monomers from the autoinhibited dimer, enables phosphorylation of Thr-402 and allows the kinase domain to adopt an active structure. Phosphorylation at Tyr, 130, Ser-141and Thr-402 increases phosphotransferase activity. Following caspase cleavage, autophosphorylated PAK-2p34 is constitutively active.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Abl1 | P00519 | S618 | APTPPKRSSSFREMD | + |
| Abl1 | P00519 | S619 | PTPPKRSSSFREMDG | + |
| Abl1 iso2 | P00519-2 | S637 | APTPPKRSSSFREMD | + |
| Abl1 iso2 | P00519-2 | S638 | PTPPKRSSSFREMDG | + |
| ARHGDIA | P52565 | S101 | LESFKKQSFVLKEGV | |
| ARHGDIA | P52565 | S34 | YKPPAQKSIQEIQEL | |
| Caldesmon | Q05682 | S714 | EGVRNIKSMWEKGNV | |
| Caldesmon | Q05682 | S744 | GLKVGVSSRINEWLT | |
| eIF4G | Q04637 | S896 | RDIARRRSLGNIKFI | |
| ERK3 (MAPK6) | Q16659 | S189 | YSHKGHLSEGLVTKW | + |
| ERK4 (MAPK4) | P31152 | S186 | YSHKGYLSEGLVTKW | + |
| Jun (c-Jun) | P05412 | T2 | ______MTAKMETTF | |
| Jun (c-Jun) | P05412 | T286 | RLEEKVKTLKAQNSE | |
| Jun (c-Jun) | P05412 | T8 | AKMETTFYDDALNAS | |
| Jun (c-Jun) | P05412 | T89 | QSSNGHITTTPTPTQ | |
| Jun (c-Jun) | P05412 | T93 | GHITTTPTPTQFLCP | + |
| MEK1 (MAP2K1) | Q02750 | S298 | RTPGRPLSSYGMDSR | + |
| Merlin | P35240 | S518 | DTDMKRLSMEIEKEK | |
| MNK1 (MKNK1) | Q9BUB5 | S39 | RRGRATDSLPGKFED | ? |
| MRLC2 (MYL12B) | P19105 | S2 | _______SSKRTKTK | + |
| MRLC2 (MYL12B) | P19105 | T19 | KKRPQRATSNVFAMF | + |
| Myc | P01106 | S373 | RRNELKRSFFALRDQ | |
| Myc | P01106 | T358 | EENVKRRTHNVLERQ | |
| Myc | P01106 | T400 | VVILKKATAYILSVQ | |
| MYLK1 (smMLCK) | Q15746 | S1208 | MKSRRPKSSLPPVLG | |
| MYLK1 (smMLCK) | Q15746 | S1759 | VRAIGRLSSMAMISG | |
| PAK2 | Q13177 | S141 | TVKQKYLSFTPPEKD | + |
| PAK2 | Q13177 | S19 | PAPPVRMSSTIFSTG | |
| PAK2 | Q13177 | S192 | PRPDHTKSIYTRSVI | |
| PAK2 | Q13177 | S197 | TKSIYTRSVIDPVPA | |
| PAK2 | Q13177 | S20 | APPVRMSSTIFSTGG | |
| PAK2 | Q13177 | S55 | KPRHKIISIFSGTEK | |
| PAK2 | Q13177 | T402 | PEQSKRSTMVGTPYW | + |
| prolactin | P01236 | S179 | NEIYPVWSGLPSLQM | |
| prolactin | P01236 | S207 | LHCLRRDSHKIDNYL | |
| SYN1 | P17600 | S551 | PAARPPASPSPQRQA | |
| SYN1 | P17600 | S605 | AGPTRQASQAGPVPR | |
| SYN1 | P17600 | S9 | NYLRRRLSDSNFMAN |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
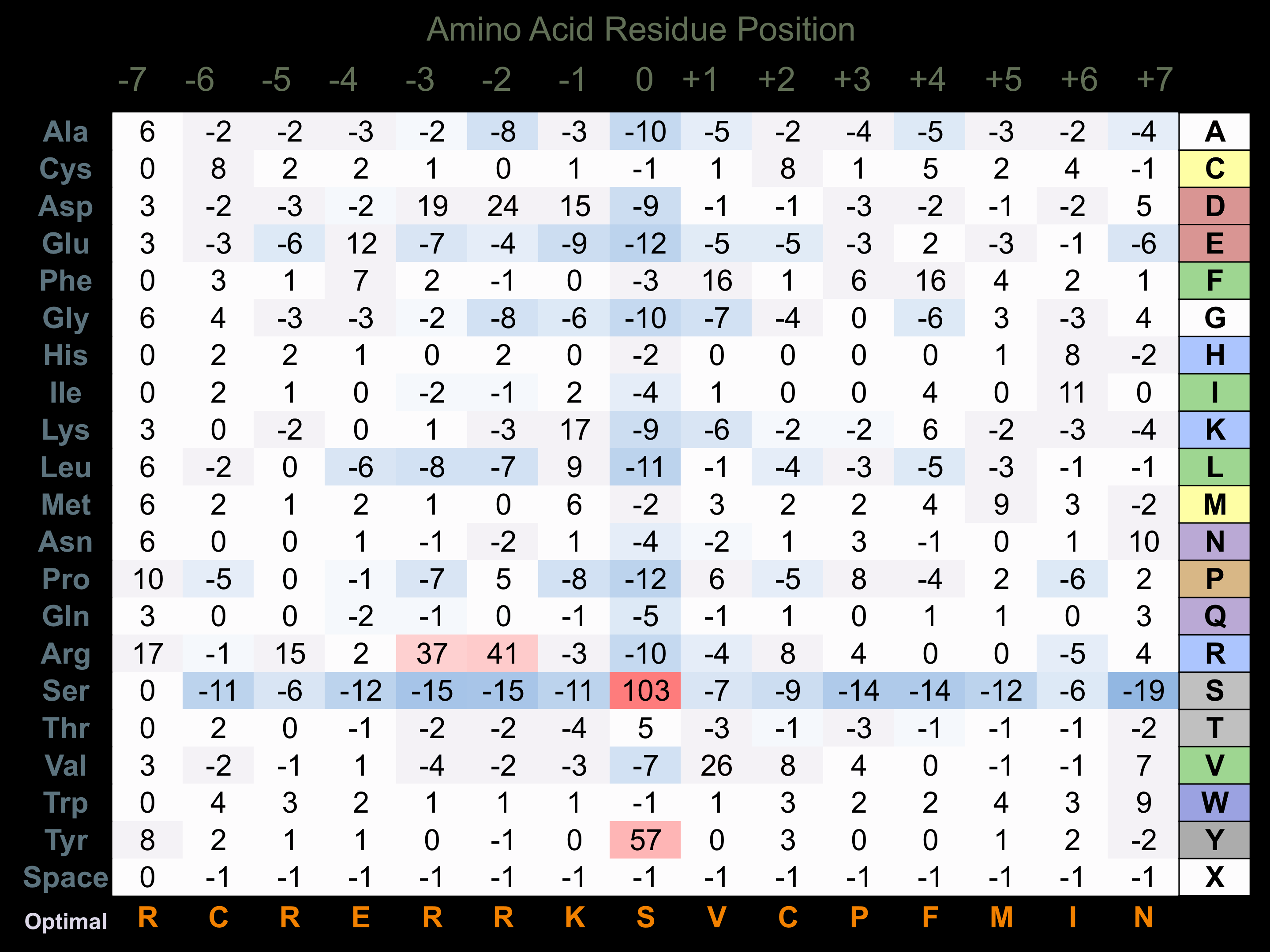
Matrix Type:
Experimentally derived from alignment of 30 known protein substrate phosphosites and 91 peptides phosphorylated by recombinant PAK2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Mental Retardation (MR); Mental Retardation, X-Linked (MRX30); Mental Retardation, X-Linked 30/47; Mental Retardation, X-Linked 17/31, Microduplication
Specific Cancer Types:
Ovarian cancer; Gastric cancer
Comments:
PAK2 may be an oncoprotein (OP). Significantly elevated S20 phosphorylated PAK2 was observed in ovarian cancer cell lines. In addition, knockdown of PAK2 expression in ovarian cancer cell lines reduces cell migration and invasion without affecting cell proliferation or apoptosis. Therefore, p-PAK2 is suggested to play an important role in ovarian carcinogenesis. In addition, elevated PAK2 and pPAK2 expression have been demonstrated in human gastric cancer cell lines. Furthermore, elevated PAK2 and pPAK2 expression was correlated with several unfavourable variables of the gastric cancer including higher tumour depth, greater extent of metastasis, and advanced tumour stage. Patients with higher expression of PAK2 and pPAK2 also displayed significantly lower survival rates than those with lower levels of expression. Therefore, PAK2 over activation may contribute to tumour progression in gastric cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +79, p<0.0007); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +72, p<0.0002); Oral squamous cell carcinomas (OSCC) (%CFC= +53, p<0.0001); and Prostate cancer - primary (%CFC= +154, p<0.0001).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24433 diverse cancer specimens. This rate is very similar (+ 6% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 6 in 20,632 cancer specimens
Comments:
Only 3 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

