Nomenclature
Short Name:
PKCa
Full Name:
Protein kinase C, alpha type
Alias:
- AAG6
- PKC III
- PKC-A
- PKC-alpha
- PKC-III; PRKCA; PRKACA; Protein kinase C, alpha; Protein kinase C, alpha type
- EC 2.7.11.13
- Kinase PKC-alpha
- KPCA
- MGC129900
- MGC129901
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKC
SubFamily:
Alpha
Specific Links
Structure
Mol. Mass (Da):
76764
# Amino Acids:
672
# mRNA Isoforms:
1
mRNA Isoforms:
76,750 Da (672 AA; P17252)
4D Structure:
Interacts with ADAP1/CENTA1, CSPG4 and PRKCABP. Binds to SDPR in the presence of phosphatidylserine. Interacts with PICK1 (via PDZ domain). Interacts with TRIM41
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
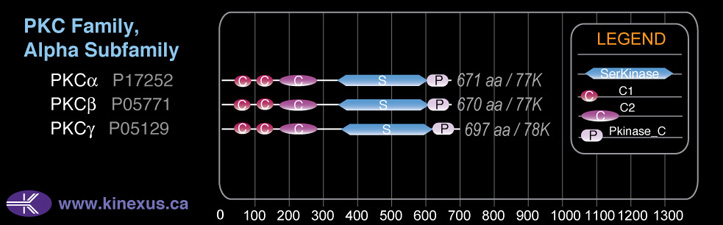
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K105, K197, K570, K604 (N6), K628 (N6).
Serine phosphorylated:
S10, S51, S208, S226, S234, S241, S319, S321, S349S405, S473, S651, S656, S657+.
Threonine phosphorylated:
T48, T176, T214, T228, T337, T361, T409, T494+, T495+, T497+, T501-, T574, T638+.
Tyrosine phosphorylated:
Y195, Y285, Y286, Y365, Y504-, Y512, Y657, Y658.
Ubiquitinated:
K628.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 30
30
853
70
1335
 6
6
178
33
254
 5
5
143
19
181
 17
17
500
218
878
 16
16
459
53
385
 17
17
487
183
1953
 9
9
265
80
461
 95
95
2742
93
5455
 16
16
465
34
448
 3
3
97
185
115
 2
2
58
63
74
 20
20
578
358
635
 2
2
57
63
79
 2
2
63
26
85
 1.4
1.4
40
36
40
 4
4
114
36
207
 4
4
101
409
1199
 3
3
85
39
194
 2
2
47
181
56
 12
12
339
241
393
 9
9
272
46
368
 2
2
60
55
78
 3
3
95
31
82
 1.3
1.3
37
37
42
 1.3
1.3
37
47
48
 100
100
2877
139
5984
 1.2
1.2
34
69
51
 3
3
78
39
181
 3
3
87
39
111
 3
3
77
56
63
 25
25
714
36
904
 93
93
2686
71
7365
 15
15
443
134
914
 27
27
778
135
690
 2
2
70
74
46
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 80.2
80.2
88.8
100 -
-
-
98 -
-
-
- 86.7
86.7
87.1
99 -
-
-
- 98.3
98.3
98.8
100 99.4
99.4
99.6
99 -
-
-
- 97.1
97.1
98.2
- 93.9
93.9
96.7
94 78.4
78.4
87
92 34.6
34.6
52.2
85 -
-
-
- 67.1
67.1
80.7
71 -
-
-
- 66.4
66.4
79.7
71 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 25.3
25.3
38.3
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | AKAP5 - P24588 |
| 2 | LMNA - P02545 |
| 3 | LMNB1 - P20700 |
| 4 | RAC1 - P63000 |
| 5 | ITGB1 - P05556 |
| 6 | GJA1 - P17302 |
| 7 | PICK1 - Q9NRD5 |
| 8 | PEBP1 - P30086 |
| 9 | ITGB4 - P16144 |
| 10 | SNAP23 - O00161 |
| 11 | PEA15 - Q15121 |
| 12 | GPM6A - P51674 |
| 13 | MYLK - Q15746 |
| 14 | GABRR2 - P28476 |
| 15 | POLB - P06746 |
Regulation
Activation:
Activated by binding of calcium, diacylglycerol and acidic phospholipids such as phosphatidylserine. Fatty acids can also activate PKCa. Phosphorylation of Thr-494 and Thr-497 increases and is essential for phosphotransferase activity. Phosphorylation of Thr-495 increases phosphotransferase activity. Phosphorylation of Ser-657 increases phosphotransferase activity, protects against dephosphorylation of the Thr-497 site and reduces the rate of degradation of PKC-alpha. Phosphorylation of Thr-638 is not essential for catalytic activity, but it protects against dephosphorylation of the Thr-497 site.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 4E-BP1 | Q13541 | S64 | FLMECRNSPVTKTPP | |
| AA-NAT | Q16613 | T31 | PSCQRRHTLPASEFR | |
| ABCB1 | P08183 | S661 | SSNDSRSSLIRKRST | |
| ABCB1 | P08183 | S667 | SSLIRKRSTRRSVRG | |
| ABCB1 | P08183 | S671 | RKRSTRRSVRGSQAQ | |
| ADAM17 | P78536 | T735 | KPFPAPQTPGRLQPA | |
| ADCY6 | O43306 | S568 | TFLILGASQKRKEEK | |
| ADCY6 | O43306 | S662 | EDLEKKYSRKVDPRF | |
| ADCY6 | O43306 | T919 | HAQQVESTARLDFLW | |
| ADD1 | P35611 | S716 | GSPGKSPSKKKKKFR | |
| ADD1 | P35611 | S726 | KKKFRTPSFLKKSKK | |
| ADD2 | P35612 | S703 | GSPSKSPSKKKKKFR | |
| ADD2 | P35612 | S713 | KKKFRTPSFLKKSKK | |
| ADD2 | P35612 | S726 | KKKEKVES_______ | |
| ADD3 | Q9UEY8 | S693 | KKKFRTPSFLKKNKK | |
| ADRA1B | P35368 | S395 | RPWTRGGSLERSQSR | |
| ADRA1B | P35368 | S401 | GSLERSQSRKDSLDD | |
| ADRA2A | P08913 | S232 | RRTRVPPSRRGPDAV | |
| ADRB2 | P07550 | S262 | GHGLRRSSKFCLKEH | |
| AFAP | Q8N556 | S277 | AELEKKLSSERPSSD | |
| AhR | P35869 | S12 | SANITYASRKRRKPV | |
| AhR | P35869 | S36 | EGIKSNPSKRHRDRL | |
| AID | Q9GZX7 | T140 | GVQIAIMTFKDYFYC | |
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| Akt1 (PKBa) | P31749 | T308 | KDGATMKTFCGTPEY | + |
| ANXA1 | P04083 | S26 | EYVQTVKSSKGGPGS | |
| ANXA1 | P04083 | S27 | YVQTVKSSKGGPGSA | |
| ANXA1 | P04083 | T23 | EEQEYVQTVKSSKGG | |
| ANXA2 | P07355 | S25 | TPPSAYGSVKAYTNF | |
| APP | P05067 | S730 | LKKKQYTSIHHGVVE | |
| AQP1 | P29972 | T156 | VLCVLATTDRRRRDL | |
| AQP1 | P29972 | T238 | APRSSDLTDRVKVWT | |
| AQP4 | P55087 | S180 | TIFASCDSKRTDVTG | |
| AR | P10275 | S578 | YGALTCGSCKVFFKR | - |
| AT2 | P50052 | S354 | MSCRKSSSLREMETF | |
| ATF2 | P15336 | S121 | LATPIIRSKIEEPSV | |
| ATP1A1 | P05023 | S16 | KYEPAAVSEQGDKKG | |
| ATP2B4 | P23634 | S1115 | NTFQTGASFKGVLRR | |
| BARK1 (GRK2, ADRBK1) | P25098 | S29 | ATPAARASKKILLPE | + |
| Basonuclin 1 | Q01954 | S541 | KKKSRKSSMPIKIEK | |
| Bcl-2 | P10415 | S70 | RDPVARTSPLQTPAA | |
| bFGF | P09038 | S206 | AEERGVVSIKGVCAN | |
| BKR2 | P30411 | S373 | SMGTLRTSISVERQI | |
| C5aR | P21730 | S314 | FQGRLRKSLPSLLRN | |
| C5aR | P21730 | S317 | RLRKSLPSLLRNVLT | |
| C5aR | P21730 | S327 | RNVLTEESVVRESKS | |
| C5aR | P21730 | S332 | EESVVRESKSFTRST | |
| C5aR | P21730 | S334 | SVVRESKSFTRSTVD | |
| C5aR | P21730 | S338 | ESKSFTRSTVDTMAQ | |
| CACNA1B | Q00975 | S773 | SVWEQRASQLRLQNL | |
| CACNA1B | Q00975 | S891 | ERPRPHRSHSKEAAG | |
| CACNA1B | Q00975 | S893 | PRPHRSHSKEAAGPP | |
| CACNA1B | Q00975 | S898 | ARPRRSHSKEAPGAD | |
| CACNA1B | Q00975 | T801 | EERLRFATTRHLRPD | |
| CACNA1C | Q13936 | S1981 | ASLGRRASFHLECLK | |
| CACNA1S | Q13698 | S1617 | NFLERTNSLPPVMAN | |
| CACNA1S | Q13698 | S687 | EKKRRKMSKGLPDKS | |
| Caldesmon | Q05682 | S643 | CFTPKGSSLKIEERA | |
| Caldesmon | Q05682 | S656 | RAEFLNKSVQKSSGV | |
| Caldesmon | Q05682 | S789 | QSVDKVTSPTKV___ | |
| Calponin 1 | P51911 | S175 | MGTNKFASQQGMTAY | |
| Calponin 1 | P51911 | T170 | IIGLQMGTNKFASQQ | |
| CaMK2a | Q9UQM7 | T286 | SCMHRQETVDCLKKF | + |
| Cannabinoid receptor 1 | P47746 | S316 | IQRGTQKSIIIHTSE | |
| CaR | P41180 | T888 | FKVAARATLRRSNVS | |
| CBP | Q92793 | S437 | CLPLKNASDKRNQQT | |
| CD3G | P09693 | S148 | GVRQSRASDKQTLLP | |
| CD4 | P01730 | S433 | RRQAERMSQIKRLLS | |
| CD4 | P01730 | S440 | SQIKRLLSEKKTCQC | |
| CD44 | P16070 | S672 | AVCIAVNSRRRCGQK | |
| CD5 | P06127 | T434 | MSFHRNHTATVRSHA | |
| CD5 | P06127 | T436 | FHRNHTATVRSHAEN | |
| CDK1 (CDC2) | P06493 | S277 | YDPAKRISGKMALNH | |
| CEACAM1 | P13688 | S508 | QQPTQPTSASPSLTA | |
| CFLAR | O15519 | S193 | LQAAIQKSLKDPSNN | |
| CFTR | P13569 | S641 | QNLQPDFSSKLMGCD | |
| CFTR | P13569 | S660 | FSAERRNSILTETLH | |
| CFTR | P13569 | S686 | WTETKKQSFKQTGEF | |
| CFTR | P13569 | S707 | SILNPINSIRKFSIV | |
| CFTR | P13569 | S737 | EPLERRLSLVPDSEQ | |
| CFTR | P13569 | S790 | IHRKTTASTRKVSLA | |
| CFTR | P13569 | S795 | TASTRKVSLAPQANL | |
| CFTR | P13569 | S813 | DIYSRRLSQETGLEI | |
| CFTR | P13569 | T604 | NKTRILVTSKMEHLK | |
| CFTR | P13569 | T682 | APVSWTETKKQSFKQ | |
| ChAT | P28329 | S464 | LLKHMTQSSRKLIRA | |
| ChAT | P28329 | S465 | LKHMTQSSRKLIRAD | |
| ChAT | P28329 | S558 | VPTYESASIRRFQEG | |
| ChAT | P28329 | S594 | HKAAVPASEKLLLLK | |
| ChAT | P28329 | T373 | TVLVKDSTNRDSLDM | |
| Cip1 (p21, CDKN1A) | P38936 | S145 | GRKRRQTSMTDFYHS | |
| Cip1 (p21, CDKN1A) | P38936 | S152 | SMTDFYHSKRRLIFS | - |
| CK1d (CSNK1D) | P48730 | S370 | MERERKVSMRLHRGA | - |
| Claudin 4 | O14493 | S194 | PRTDKPYSAKYSAAR | |
| Connexin 32 | P08034 | S233 | NPPSRKGSGFGHRLS | |
| Coronin 1B | Q9BR76 | S2 | ______MSFRKVVRQ | |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| CREM | Q03060-2 | S71 | EILSRRPSYRKILNE | |
| CTNND1 | O60716 | S879 | LIDRNQKSDKKPDRE | |
| CTPS | P17812 | S462 | TLFQTKNSVMRKLYG | |
| CTPS | P17812 | T455 | MRLGKRRTLFQTKNS | |
| Cx43 | P17302 | S261 | SPAKDCGSQKYAYFN | |
| Cx43 | P17302 | S363 | AIVDQRPSSRASSRA | |
| Cx43 | P17302 | S364 | IVDQRPSSRASSRAS | |
| Cx43 | P17302 | S367 | QRPSSRASSRASSRP | |
| Cx43 | P17302 | S368 | RPSSRASSRASSRPR | |
| Cx43 | P17302 | S371 | SRASSRASSRPRPDD | |
| Cx43 | P17302 | S372 | RASSRASSRPRPDDL | |
| Cytohesin 2 | Q99418 | S392 | AARKKRISVKKKQEQ | |
| DAB2 | P98082 | S24 | QAAPKAPSKKEKKKG | |
| Desmin | P17661 | S12 | SSSQRVSSYRRTFGG | |
| Desmin | P17661 | S47 | GSKGSSSSVTSRVYQ | |
| Desmin | P17661 | S67 | GGAGGLGSLRASRLG | |
| DGK-D | Q16760 | S66 | MLTKQNNSFQRSKRR | |
| DGK-D | Q16760 | S70 | QNNSFQRSKRRYFKL | |
| DGK-Z | Q13574 | S447 | PQNTLKASKKKKRAS | - |
| DGK-Z | Q13574 | S454 | SKKKKRASFKRKSSK | - |
| DGK-Z | Q13574 | S459 | RASFKRKSSKKGPEE | - |
| DGK-Z | Q13574 | S460 | ASFKRKSSKKGPEEG | - |
| Dlx3 | O60479 | S138 | KPRTIYSSYQLAALQ | |
| DNAM1 | Q15762 | S329 | YVNYPTFSRRPKTRV | |
| DOR1 | P41143 | S344 | CGRPDPSSFSRAREA | |
| DYN1 | Q05193 | S795 | VPPARPGSRGPAPGP | |
| EGFR | P00533 | T678 | RHIVRKRTLRRLLQE | - |
| EGLN2 | Q96KS0 | S132 | GGDAPSPSKRPWARQ | |
| EGLN2 | Q96KS0 | S226 | LRDGQLVSQRAIPPR | |
| EGLN2 | Q96KS0 | S234 | QRAIPPRSIRGDQIA | |
| eIF2B | P20042 | S13 | MIFDPTMSKKKKKKK | |
| eIF4E | P06730 | S209 | DTATKSGSTTKNRFV | + |
| eIF4E | P06730 | T210 | TATKSGSTTKNRFVV | |
| eIF6 | P56537 | S235 | QPSTIATSMRDSLID | |
| ELAVL1 | Q15717 | S158 | FIRFDKRSEAEEAIT | |
| ELAVL1 | Q15717 | S221 | QAQRFRFSPMGVDHM | |
| eNOS | P29474 | T495 | TGITRKKTFKEVANA | |
| EP300 | Q09472 | S89 | SELLRSGSSPNLNMG | - |
| ErbB2 (HER2) | P04626 | T686 | QQKIRKYTMRRLLQE | - |
| Ezrin | P15311 | T567 | QGRDKYKTLRQIRQG | |
| F11R | Q9Y624 | S284 | KVIYSQPSARSEGEF | |
| F3 | P13726 | S285 | RKAGVGQSWKENSPL | |
| F3 | P13726 | S290 | GQSWKENSPLNVS__ | |
| Fascin | Q61558 | S38 | KVNASASSLKKKQIW | |
| FMIP | Q13769 | S5 | ___MSSESSKKRKPK | |
| FMIP | Q13769 | S6 | __MSSESSKKRKPKV | |
| FRAT1 | Q92837 | S188 | RLQQRRGSQPETRTG | |
| FSH receptor | P23945 | T470 | ITLERWHTITHAMQL | |
| FSH receptor | P23945 | T472 | LERWHTITHAMQLDC | |
| Ga(z) | P19086 | S16 | EKEAARRSRRIDRHL | |
| Ga(z) | P19086 | S27 | DRHLRSESQRQRREI | |
| Ga(z) | P19086 | Y155 | HLEDNAAYYLNDLER | |
| GABRB1 | P18505 | S434 | GRIRRRASQLKVKIP | |
| GABRB3 | P28472 | S432 | KTHLRRRSSQLKIKI | |
| GABRG2 | P18507 | S433 | THLRRRSSQLKIKIP | |
| GAP43 | P17677 | S41 | AATKIQASFRGHITR | |
| GBPI-1 | Q9NXH3 | T58 | SRRPSRLTVKYDRGQ | |
| GFAP | P14136 | S13 | ITSAARRSYVSSGEM | |
| GFAP | P14136 | S38 | LGPGTRLSLARMPPP | |
| GFAP | P14136 | S8 | MERRRITSAARRSYV | |
| Gg(12) | Q9UBI6 | S2 | ______MSSKTASTN | |
| Gg(12) | Q9UBI6 | S3 | _____MSSKTASTNN | |
| GIRK1 | P48549 | S185 | GCMFIKMSQPKKRAE | |
| GIRK4 | P48544 | S191 | GCMFVKISQPKKRAE | |
| GLP1R | P43220 | S431 | HLHIQRDSSMKPLKC | |
| GLP1R | P43220 | S432 | LHIQRDSSMKPLKCP | |
| GluR-d2 | O43424 | S945 | QTLSRTLSAKAASGF | |
| GluR1 | P42261 | S836 | CYKSRSESKRMKGFC | |
| GluR1 | P42261 | S849 | FCLIPQQSINEAIRT | |
| GluR1 | P42261 | T858 | NEAIRTSTLPRNSGA | |
| GluR2 | P42262 | S863 | NPSSSQNSQNFATYK | |
| GluR2 | P42262 | S880 | YNVYGIESVKI____ | |
| GluR4 | P48058 | S862 | IRNKARLSITGSVGE | |
| GluR5 | P39086-2 | S879 | QKKIKKKSRTKGKSS | |
| GluR5 | P39086-2 | S885 | KSRTKGKSSFTSILT | |
| GMFb | P60983 | S71 | QPRFIVYSYKYQHDD | |
| GRK7 | Q8WTQ7 | S36 | ELQRRRRSLALPGLQ | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| GSTA4 | O15217 | T193 | VKLSNIPTIKRFLEP | |
| GSTP1 | P09211 | S184 | SAYVGRLSARPKLKA | |
| H-Ras-1 | P01112 | S177 | KLNPPDESGPGCMSC | |
| H11 (HSPB8) | Q9UJY1 | S14 | PFSCHYPSRLRRDPF | |
| H11 (HSPB8) | Q9UJY1 | T63 | LSSAWPGTLRSGMVP | |
| H1D | P16403 | S104 | KGTGASGSFKLNKKA | |
| H1D | P16403 | S36 | GGTPRKASGPPVSEL | |
| H1R | P35367 | S396 | FTWKRLRSHSRQYVS | |
| H1R | P35367 | S398 | WKRLRSHSRQYVSGL | |
| H1R | P35367 | T478 | CNENFKKTFKRILHI | |
| H2B | P33778 | S33 | DGKKRKRSRKESYSI | |
| H2B | P33778 | S37 | RKRSRKESYSIYVYK | |
| HIR | P48050 | T53 | IFTTCVDTRWRYMLM | |
| HMG-CoA reductase | P04035 | S872 | SHMIHNRSKINLQDL | |
| HMGA1 | P17096 | S44 | PGTALVGSQKEPSEV | |
| HMGA1 | P17096 | S64 | PRGRPKGSKNKGAAK | |
| hnRNP A1 | P09651 | S192 | EMASASSSQRGRSGS | |
| hnRNP A1 | P09651 | S197 | SSSQRGRSGSGNFGG | |
| hnRNP A1 | P09651 | S199 | SQRGRSGSGNFGGGR | - |
| hnRNP A1 | P09651 | S223 | FGRGGNFSGRGGFGG | |
| hnRNP A1 | P09651 | S231 | GRGGFGGSRGGGGYG | |
| hnRNP A1 | P09651 | S95 | RAVSREDSQRPGAHL | |
| hnRNP A1 | P09651 | T103 | QRPGAHLTVKKIFVG | |
| HOXA9 | P31269 | S205 | ANWLHARSTRKKRCP | - |
| HOXA9 | P31269 | T206 | NWLHARSTRKKRCPY | - |
| HSER (GuanyCyclase) | P25092 | S1052 | QKPRRVASYKKGTLE | + |
| HSP27 | P04792 | S15 | FSLLRGPSWDPFRDW | ? |
| HSP27 | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| ICLN | P54105 | S45 | YIAESRLSWLDGSGL | |
| IKKb (IKBKINASE) | O14920 | S177 | AKELDQGSLCTSFVG | + |
| IKKb (IKBKINASE) | O14920 | S181 | DQGSLCTSFVGTLQY | + |
| IL2RA | P01589 | S268 | WQRRQRKSRRTI___ | |
| IL2RA | P01589 | T271 | RQRKSRRTI______ | |
| InsR | P06213 | S1062 | AVKTVNESASLRERI | |
| InsR | P06213 | S1064 | KTVNESASLRERIEF | |
| InsR | P06213 | T1375 | KKNGRILTLPRSNPS | + |
| IREB1 | P21399 | S138 | DFNRRADSLQKNQDL | |
| IREB1 | P21399 | S711 | REFNSYGSRRGNDAV | |
| IRS1 | P35568 | S24 | GYLRKPKSMHKRFFV | |
| IRS1 | P35568 | S312 | TESITATSPASMVGG | - |
| IRS1 | P35568 | S323 | MVGGKPGSFRVRASS | |
| IRS1 | P35568 | S616 | DDGYMPMSPGVAPVP | - |
| ITGB2 | P05107 | S745 | FEKEKLKSQWNNDNP | |
| ITGB2 | P05107 | T758 | NPLFKSATTTVMNPK | |
| ITGB2 | P05107 | T760 | LFKSATTTVMNPKFA | |
| ITGB4 | P16144 | S1356 | YSDDVLRSPSGSQRP | |
| ITGB4 | P16144 | S1364 | PSGSQRPSVSDDTGC | |
| ITGB4 | P16144 | S1494 | TLTRDYNSLTRSEHS | |
| ITGB4 | P16144 | S1360 | VLRSPSGSQRPSVSD | |
| KCNAB2 (Kvb2) | Q13303 | S266 | IPPYSRASLKGYQWL | |
| KCNE3 | Q9Y6H6 | S82 | LILGYTRSRKVDKRS | |
| KCNJ13 | O60928 | S201 | TRPSPLTSVRVSAVL | |
| KCNJ8 | Q15842 | S379 | SELSHQNSLRKRNSM | |
| KCNJ8 | Q15842 | S385 | NSLRKRNSMRRNNSM | |
| KCNJ8 | Q15842 | S391 | NSMRRNNSMRRNNSI | |
| KCNJ8 | Q15842 | S397 | NSMRRNNSIRRNNSS | |
| KIR3DL1 | P43629 | S415 | QRKITRPSQRPKTPP | |
| Kir6.2 | Q14654 | T180 | QAHRRAETLIFSKHA | |
| Kit | P10721 | S741 | TKADKRRSVRIGSYI | - |
| Kit | P10721 | S746 | RRSVRIGSYIERDVT | - |
| KRT20 | P35900 | S13 | RSFHRSLSSSLQAPV | |
| Kv1.5 | P22460 | T15 | LENGGAMTVRGGDEA | - |
| Kv3.4 | Q03721 | S15 | SSYRGRKSGNKPPSK | |
| Kv3.4 | Q03721 | S21 | KSGNKPPSKTCLKEE | |
| Kv3.4 | Q03721 | S8 | MISSVCVSSYRGRKS | |
| Kv3.4 | Q03721 | S9 | ISSVCVSSYRGRKSG | |
| Kv4.2 | Q9NZV8 | S447 | SANAYMQSKRNGLLS | |
| Kv4.2 | Q9NZV8 | S537 | CSRRHKKSFRIPNAN | |
| Lamin A,C | P48678 | S5 | ___METPSQRRATRS | |
| Lamin A,C | P02545 | S525 | NTWGCGNSLRTALIN | |
| Lamin A,C | P02545 | S573 | HGSHCSSSGDPAEYN | |
| Lamin A,C | P02545 | S651 | RSYLLGNSSPRTQSP | |
| Lamin A,C | P02545 | T199 | DAENRLQTMKEELDF | |
| Lamin A,C | P02545 | T416 | TQGGGSVTKKRKLES | |
| Lamin A,C | P02545 | T480 | NGDDPLLTYRFPPKF | |
| Lck | P06239 | S42 | TLLIRNGSEVRDPLV | |
| MAP4 | P27816 | S868 | PKSTSTSSMKKTTTL | |
| MARCKS | P29966 | S159 | KKKKKRFSFKKSFKL | |
| MARCKS | P29966 | S163 | KRFSFKKSFKLSGFS | |
| MARCKS | P29966 | S167 | FKKSFKLSGFSFKKN | |
| MARCKS | P29966 | S170 | SFKLSGFSFKKNKKE | |
| MAT1A | Q00266 | T341 | TYGTSQKTERELLDV | |
| Mena iso4 | Q8N8S7 | S265 | WERERRISSAAAPAS | |
| Met | P08581 | S985 | PHLDRLVSARSVSPT | - |
| mGluR5 | P41594 | S840 | VRSAFTTSTVVRMHV | |
| mGluR5 iso2 | P41594 | S934 | QHLWQRLSIHINKKE | |
| mGluR7 | Q14831 | S862 | NVQKRKRSFKAVVTA | |
| MKLN1 | Q9UL63 | S324 | TEKENGPSARSCHKM | |
| MKLN1 | Q9UL63 | T515 | MVPMTGFTQRATIDP | |
| MRLC1 (MYL9) | P24844 | S2 | ______MSSKRAKAK | |
| MRLC1 (MYL9) | P24844 | S20 | KRPQRATSNVFAMFD | |
| MRLC1 (MYL9) | P24844 | S3 | _____MSSKRAKAKT | |
| MRLC1 (MYL9) | P24844 | T10 | SKRAKAKTTKKRPQR | |
| MT1 | P04731 | S32 | KCNSCKKSCCSCCPM | |
| MUNC18 (STXBP1) | P61764 | S306 | VSQEVTRSLKDFSSS | |
| MUNC18 (STXBP1) | P61764 | S313 | SLKDFSSSKRMNTGE | |
| Myelin P0 | P25189 | S210 | HKPGKDASKRGRQTP | |
| Myelin P0 | P25189 | S233 | SRSTKAVSEKKAKGL | |
| Myelin P0 | P25189 | S243 | KAKGLGESRKDKK__ | |
| Myf-6 | P23409 | S220 | SSSLRCLSSIVDSIS | |
| Myf-6 | P23409 | S221 | SSLRCLSSIVDSISS | |
| Myf-6 | P23409 | T99 | TDRRKAATLRERRRL | |
| MYH9 | P35579 | S1915 | AMNREVSSLKNKLRR | |
| Myogenin | P15173 | T87 | VDRRRAATLREKRRL | |
| Neurogranin | Q92686 | S36 | AAAKIQASFRGHMAR | |
| NFE2L2 | Q16236 | S40 | SREVFDFSQRRKEYE | |
| NFkB-p105 | P19838 | S337 | FVQLRRKSDLETSEP | |
| NFkB-p65 | Q04206 | S276 | SMQLRRPSDRELSEP | |
| NFL (Neurofilament L) | P07196 | S13 | YEPYYSTSYKRRYVE | |
| NFL (Neurofilament L) | P07196 | S28 | TPRVHISSVRSGYST | |
| NFL (Neurofilament L) | P07196 | S34 | SSVRSGYSTARSAYS | |
| NFL (Neurofilament L) | P07196 | S52 | APVSSSLSVRRSYSS | |
| NG2 | Q6UVK1 | T2252 | YLRKRNKTGKHDVQV | |
| NHERF | O14745 | S162 | CTMKKGPSGYGFNLH | |
| NHERF | O14745 | S339 | ERAHQKRSSKRAPQM | |
| NHERF | O14745 | S340 | RAHQKRSSKRAPQMD | |
| NHERF | O14745 | S76 | ETHQQVVSRIRAALN | |
| NKX3-1 | Q99801 | S48 | RQGGRTSSQRQRDPE | - |
| NMDAR1 | Q05586 | S889 | AITSTLASSFKRRRS | |
| NMDAR1 | Q05586 | S897 | SFKRRRSSKDTSTGG | |
| NMDAR1 (Glutamate) | Q05586 | S890 | ITSTLASSFKRRRSS | |
| NMDAR1 (Glutamate) | Q05586 | S896 | SSFKRRRSSKDTSTG | |
| NMDAR2A | Q12879 | S1416 | ASYCSRDSRGHNDVY | |
| NMDAR2B (GRIN2B) | Q13224 | S1303 | NKLRRQHSYDTFVDL | - |
| NMDAR2B (GRIN2B) | Q13224 | S1323 | ALAPRSVSLKDKGRF | |
| NMDAR2C | Q14957 | S1227 | PCTWRRISSLESEV | |
| NMNAT1 | Q9HAN9 | S136 | WTETQDSSQKKSLEP | |
| NPHS1 | O60500 | T1120 | EYEESQWTGERDTQS | + |
| NPHS1 | O60500 | T1125 | QWTGERDTQSSTVST | + |
| NR1H4 iso2 | Q96RI1-2 | S135 | VVCGDRASGYHYNAL | |
| NR1H4 iso2 | Q96RI1-2 | S154 | CKGFFRRSITKNAVY | |
| NR2C1 | P13056 | S197 | IEVSREKSSNCAAST | + |
| NR2C1 | P13056 | S568 | IGNVRIDSVIPHILK | |
| Occludin | Q16625 | S340 | DKRFYPESSYKSTPV | |
| p40phox | Q15080 | S315 | RQARGLPSQKRLFPW | |
| p40phox | Q15080 | T154 | LRRLRPRTRKVKSVS | |
| p47phox | P14598 | S303 | RGAPPRRSSIRNAHS | + |
| p47phox | P14598 | S304 | GAPPRRSSIRNAHSI | + |
| p47phox | P14598 | S315 | AHSIHQRSRKRLSQD | ? |
| p47phox | P14598 | S320 | QRSRKRLSQDAYRRN | ? |
| p47phox | P14598 | S328 | QDAYRRNSVRFLQQR | + |
| p47phox | P14598 | S359 | EERQTQRSKPQPAVP | + |
| p47phox | P14598 | S370 | PAVPPRPSADLILNR | + |
| p47phox | P14598 | S379 | DLILNRCSESTKRKL | ? |
| p53 | P04637 | S371 | AHSSHLKSKKGQSTS | + |
| p53 | P04637 | S376 | LKSKKGQSTSRHKKL | + |
| p53 | P04637 | S378 | SKKGQSTSRHKKLMF | + |
| p53 | P04637 | T377 | KSKKGQSTSRHKKLM | |
| p70S6Kb (RPS6KB2) | Q9UBS0 | S473 | PPSGTKKSKRGRGRP | + |
| p73 | O15350 | S388 | VPQPLVDSYRQQQQL | |
| PA2G4 | Q9UQ80 | S360 | ELKALLQSSASRKTQ | ? |
| PAM | P19021 | S934 | FASRKGYSRKGFDRL | |
| PDE3A | Q14432 | S312 | SKSHRRTSLPCIPRE | |
| PDE3A | Q14432 | S428 | IPKRLRRSLPPGLLR | |
| PDE3A | Q14432 | S438 | PGLLRRVSSTWTTTT | |
| PDE3A | Q14432 | S465 | VRRDRSTSIKLQEAP | |
| PDE3A | Q14432 | S492 | MTLTKSRSFTSSYAI | |
| PDE5A | O76074 | S102 | GTPTRKISASEFDRP | + |
| PEA-15 | Q15121 | S104 | TKLTRIPSAKKYKDI | |
| PEBP1 | P30086 | S153 | RGKFKVASFRKKYEL | |
| PFKFB2 | O60825 | T475 | TPLSSSNTIRRPRNY | |
| PFKFB3 | Q16875 | S461 | NPLMRRNSVTPLASP | |
| PITPNA | Q00169 | S165 | EDPAKFKSIKTGRGP | |
| PITPNA | Q00169 | T58 | DGEKGQYTHKIYHLQ | |
| PITPNB | P48739 | S164 | EDPALFQSVKTKRGP | |
| PITPNB | P48739 | S261 | ETMRKRGSVRGTSAA | |
| PKCa (PRKCA) | P17252 | S656 | QSDFEGFSYVNPQFV | |
| PKCa (PRKCA) | P17252 | T497 | MDGVTTRTFCGTPDY | + |
| PKCa (PRKCA) | P17252 | Y657 | SDFEGFSYVNPQFVH | |
| PKCb (PRKCB) | P05771 | T500 | WDGVTTKTFCGTPDY | + |
| PKCe (PRKCE) | Q02156 | S234 | ETPDQVGSQRFSVNM | |
| PKCe (PRKCE) | Q02156 | S316 | TPDKITNSGQRRKKL | |
| PKCe (PRKCE) | Q02156 | S368 | NNIRKALSFDNRGEE | ? |
| PKD1 (PRKCM) | Q15139 | S249 | GREKRSNSQSYIGRP | + |
| PKD1 (PRKCM) | Q15139 | S738 | ARIIGEKSFRRSVVG | + |
| PKD1 (PRKCM) | Q15139 | S742 | GEKSFRRSVVGTPAY | + |
| PKD2 (PRKD2) | Q9BZL6 | S706 | ARIIGEKSFRRSVVG | + |
| PKD2 (PRKD2) | Q9BZL6 | S710 | GEKSFRRSVVGTPAY | + |
| PKD2 (PRKD2) | Q9BZL6 | S876 | QGLAERISVL_____ | + |
| PKG1 (PRKG1) | Q13976 | T59 | THIGPRTTRAQGISA | + |
| PLCB1 | Q9NQ66 | S887 | HSQPAPGSVKAPAKT | |
| PLCB3 | Q01970 | S1105 | LDRKRHNSISEAKMR | - |
| PLCG1 | P19174 | S1248 | HGRAREGSFESRYQQ | |
| PLD1 iso2 | Q13393 | S2 | ______MSLKNEPRV | |
| PLD1 iso2 | Q13393 | S561 | PRKFSKFSLYKQLHR | |
| PLD1 iso2 | Q13393 | T147 | PIPTRRHTFRRQNVR | |
| PLD2 | O14939 | S243 | RWLVVKDSFLLYMCL | |
| PLD2 | O14939 | T252 | LLYMCLETGAISFVQ | |
| Pleckstrin | P08567 | S113 | GQKFARKSTRRSIRL | |
| Pleckstrin | P08567 | S117 | ARKSTRRSIRLPETI | |
| PLM | O00168 | S83 | EEGTFRSSIRRLSTR | |
| PLM | O00168 | S88 | RSSIRRLSTRRR___ | |
| PLM | O00168 | T89 | SSIRRLSTRRR____ | |
| PPAR-binding protein | Q15648 | T796 | EASKLPSTSDDCPAI | |
| PPARA | Q07869 | S142 | VYDKCDRSCKIQKKN | + |
| PPARA | Q07869 | S179 | RFGRMPRSEKAKLKA | ? |
| PPARA | Q07869 | S230 | VKARVILSGKASNNP | |
| PPARA | Q07869 | T129 | CKGFFRRTIRLKLVY | + |
| PPP1CA | P62138 | S325 | PITPPRNSAKAKK__ | |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PPP1R1A | Q13522 | S67 | LKSTLAMSPRQRKKM | |
| PPP1R1A | Q13522 | T75 | PRQRKKMTRITPTMK | + |
| PRKD3 | O94806 | S731 | ARIIGEKSFRRSVVG | + |
| PSEN1 | P49768 | S346 | EWEAQRDSHLGPHRS | |
| PTGIR | P43119 | S328 | TPLSQLASGRRDPRA | |
| PTP1B | P18031 | S378 | RSRVVGGSLRGAQAA | |
| PTPN11 (SHP2) | Q06124 | S580 | CAEMREDSARVYENV | |
| PTPN11 (SHP2) | Q06124 | S595 | GLMQQQKSFR_____ | |
| PTPN12 (PTP-PEST) | Q05209 | S39 | FMRLRRLSTKYRTEK | - |
| PTPN12 (PTP-PEST) | Q05209 | S435 | KKLERNLSFEIKKVP | |
| PTPN6 (SHP1) | P29350 | S591 | DKEKSKGSLKRK___ | |
| PTPRA | P18433 | S180 | QAGSHSNSFRLSNGR | |
| PTPRA | P18433 | S213 | PLLARSPSTNRKYPP | + |
| Rabphilin 3A | Q9Y2J0 | S237 | GPPVRRASEARMSSS | |
| Rabphilin 3A | Q9Y2J0 | S272 | AGLRRANSVQASRPA | |
| Raf1 | P04049 | S43 | FGYQRRASDDGKLTD | - |
| Raf1 | P04049 | S497 | ATVKSRWSGSQQVEQ | + |
| Raf1 | P04049 | S499 | VKSRWSGSQQVEQPT | + |
| Raf1 | P04049 | S619 | SLPKINRSASEPSLH | + |
| RALBP1 | Q15311 | S118 | VFKKPSFSKKKEKDF | |
| RALBP1 | Q15311 | S353 | LSPTVQISNRVLYVF | |
| RALBP1 | Q15311 | S509 | FLRRQIASEKEEIER | |
| RALBP1 | Q15311 | T297 | ACGRTTETEKVQEFQ | |
| RGS7 | P49802 | S434 | YKLMKSDSYPRFIRS | |
| RGS9 | O75916-3 | S478 | MSKLDRRSQLKKELP | |
| Rhodopsin | P08100 | S334 | PLGDDEASATVSKTE | |
| Rhodopsin | P08100 | S338 | DEASATVSKTETSQV | |
| ROMK | P48048 | S201 | FSKNAVISKRGGKLC | |
| ROMK | P48048 | S4 | ____MNASSRNVFDT | |
| RRAD | P55042 | S214 | LVRSREVSVDEGRAC | |
| RRAD | P55042 | S257 | QIRLRRDSKEANARR | |
| RRAD | P55042 | S273 | AGTRRRESLGKKAKR | |
| RRAD | P55042 | S290 | GRIVARNSRKMAFRA | |
| RRAD | P55042 | S299 | KMAFRAKSKSCHDLS | |
| RSK1 (RPS6KA1) | Q15418 | S363 | TSRTPKDSPGIPPSA | + |
| SCN2A | Q99250 | S1506 | NAMKKLGSKKPQKPI | |
| SCN2A | Q99250 | S576 | PRRNSRASLFNFKGR | |
| SCN2A | Q99250 | S610 | DNDSRRDSLFVPHRH | |
| SH3BP2 (3BP2) | P78314 | S225 | SDMPRAHSFTSKGPG | |
| SH3BP2 (3BP2) | P78314 | S278 | PATPRRMSDPPLSTM | |
| sialophorin | P16150 | S351 | TFFGRRKSRQGSLAM | |
| SIK | P57059 | S577 | FQEGRRASDTSLTQG | |
| SLC12A5 | Q9H2X9 | S963 | ITDESRGSIRRKNPA | |
| SLC22A1 | O15245 | S285 | YYWCVPESPRWLLSQ | |
| SLC22A1 | O15245 | S291 | ESPRWLLSQKRNTEA | |
| SLC22A1 | O15245 | T327 | LSLEEDVTEKLSPSF | |
| SLC22A1 | O15245 | T548 | TIYLKVQTSEPSGT_ | |
| SNAP23 | O00161 | S95 | NRTKNFESGKAYKTT | |
| SNAP25 | P60880 | S187 | RIMEKADSNKTRIDE | |
| SNAP25 | P60880 | T138 | GGFIRRVTNDARENE | |
| Src | P12931 | S12 | KSKPKDASQRRRSLE | + |
| SRF | P11831 | S162 | LRRYTTFSKRKTGIM | |
| SRF | P11831 | T159 | DNKLRRYTTFSKRKT | |
| SSTR2 | P30874 | S343 | DDGERSDSKQDKSRL | |
| Syndecan-2 | P34741 | S187 | DLGERKPSSAAYQKA | |
| Syndecan-2 | P34741 | S188 | LGERKPSSAAYQKAP | |
| syndecan-4 | P31431 | S179 | MKKKDEGSYDLGKKP | |
| SYT1 | P21579 | T113 | DVKDLGKTMKDQALK | |
| TACSTD2 | P09758 | S303 | VITNRRKSGKYKKVE | |
| TAGLN | Q01995 | S181 | VIGLQMGSNRGASQA | |
| Tau iso8 | P10636-8 | S258 | PDLKNVKSKIGSTEN | |
| Tau iso8 | P10636-8 | S293 | NVQSKCGSKDNIKHV | |
| Tau iso8 | P10636-8 | S305 | KHVPGGGSVQIVYKP | |
| Tau iso8 | P10636-8 | S324 | KVTSKCGSLGNIHHK | |
| Tau iso8 | P10636-8 | S352 | DFKDRVQSKIGSLDN | |
| TAUT | P31641 | S322 | YNKYKYNSYRDCMLL | |
| TBXA2R | P21731 | S324 | RRLQPRLSTRPRSLS | |
| TBXA2R | P21731 | S329 | RLSTRPRSLSLQPQL | |
| TBXA2R | P21731 | S331 | STRPRSLSLQPQLTQ | |
| TBXA2R | P21731 | T337 | LSLQPQLTQRSGLQ_ | |
| TBXA2R iso2 | P21731-2 | T399 | LLPFEPPTGKALSRK | |
| TFRC | P02786 | S24 | PLSYTRFSLARQVDG | |
| TH | P07101 | S70 | RFIGRRQSLIEDARK | |
| TMPO | P42167 | S180 | RQNGSNDSDRYSDNE | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TNNI3 | P19429 | S41 | AKKKSKISASRKLQL | |
| TNNI3 | P19429 | S43 | KKSKISASRKLQLKT | |
| TNNI3 | P19429 | S76 | GEKGRALSTRCQPLE | |
| TNNI3 | P19429 | T142 | RGKFKRPTLRRVRIS | |
| TNNT2 | P45379 | S207 | QAQTERKSGKRQTER | |
| TNNT2 | P45379 | S284 | INDNQKVSKTRGKAK | |
| TNNT2 | P45379 | T203 | YIQKQAQTERKSGKR | |
| TNNT2 | P45379 | T212 | RKSGKRQTEREKKKK | |
| TNNT2 | P45379 | T293 | TRGKAKVTGRWK___ | |
| TOP1 | P11387 | S21 | ADFRLNDSHKHKDKH | |
| TREK-2 | P57789 | S326 | GDWLRVLSKKTKEEV | |
| TREK-2 | P57789 | S359 | RETRRRLSVEIHDKL | |
| TRPC3 | Q13507 | S712 | PPFSLVPSPKSFVYF | |
| TrpC6 iso2 | Q9Y210 | S769 | VPFNLVPSPKSLFYL | |
| TRPV5 | Q9NQA5 | S299 | FLELVVSSDKREARQ | |
| TRPV5 | Q9NQA5 | S654 | YVEVFKNSDKEDDQE | |
| TUBA1C | Q9BQE3 | S158 | SLLMERLSVDYGKKS | |
| TUBA1C | Q9BQE3 | S165 | SVDYGKKSKLEFSIY | |
| uPA | P00749 | S158 | CADGKKPSSPPEELK | |
| uPA | P00749 | S323 | TGFGKENSTDYLYPE | |
| VAChT | Q16572 | S480 | GLLTRSRSERDVLLD | |
| VASP | P50552 | S154 | EHLERRVSNAGGPPA | |
| VDR | P11473 | S51 | CKGFFRRSMKRKALF | + |
| Vimentin | P08670 | S10 | TRSVSSSSYRRMFGG | |
| Vimentin | P08670 | S25 | PGTASRPSSSRSYVT | |
| Vimentin | P08670 | S26 | GTASRPSSSRSYVTT | |
| Vimentin | P08670 | S34 | SRSYVTTSTRTYSLG | |
| Vimentin | P08670 | S39 | TTSTRTYSLGSALRP | - |
| Vimentin | P08670 | S42 | TRTYSLGSALRPSTS | |
| Vimentin | P08670 | S5 | ___MSTRSVSSSSYR | |
| Vimentin | P08670 | S51 | LRPSTSRSLYASSPG | |
| Vimentin | P08670 | S66 | GVYATRSSAVRLRSS | |
| Vimentin | P08670 | S7 | _MSTRSVSSSSYRRM | |
| Vimentin | P08670 | S8 | MSTRSVSSSSYRRMF | |
| Vimentin | P08670 | S9 | STRSVSSSSYRRMFG | |
| Vinculin | P18206 | S1100 | NAQNLMQSVKETVRE | |
| Vinculin | P18206 | S1112 | VREAEAASIKIRTDA | |
| VR1 | Q8NER1 | S502 | YFLQRRPSMKTLFVD | |
| VR1 | Q8NER1 | S775 | EGVKRTLSFSLRSSR | |
| VR1 | Q8NER1 | S801 | VPLLREASARDRQSA | |
| VR1 | Q8NER1 | S821 | YLRQFSGSLKPEDAE | |
| VR1 | Q8NER1 | T145 | QKSKKHLTDNEFKDP | |
| VR1 | Q8NER1 | T705 | WKLQRAITILDTEKS | |
| VTN | P04004 | S381 | RNRKGYRSQRGHSRG | |
| VTN | P04004 | S386 | YRSQRGHSRGRNQNS | |
| VTN | P04004 | S393 | SRGRNQNSRRPSRAT | |
| VTN | P04004 | S397 | NQNSRRPSRATWLSL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
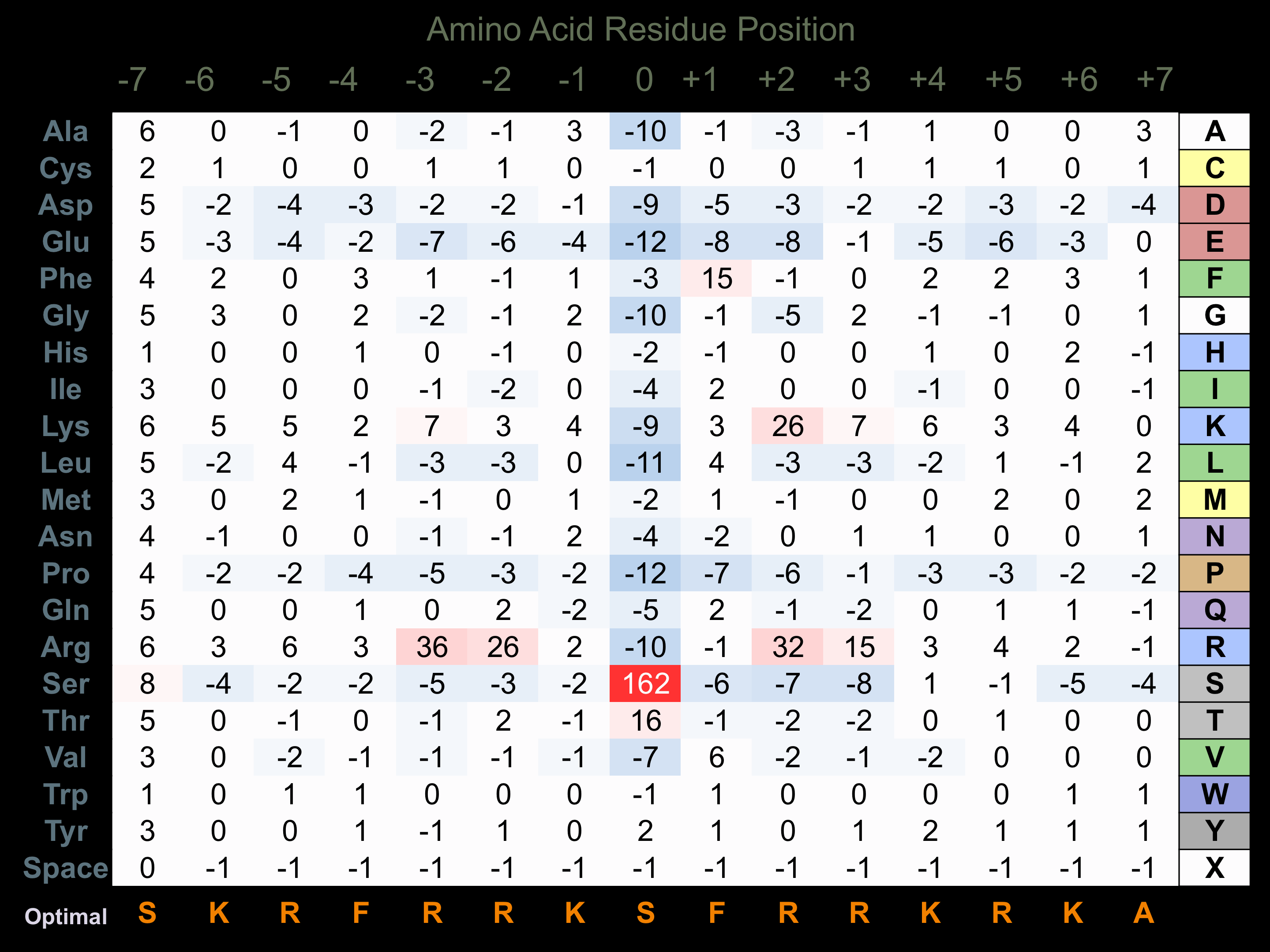
Matrix Type:
Experimentally derived from alignment of 784 known protein substrate phosphosites and 98 peptides phosphorylated by recombinant PKCa in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Comments:
Variations in PKCa’s gene sequence may also associate with increased susceptibility to asthma.
Specific Cancer Types:
Pituitary tumours, invasive
Comments:
PKCa may be an oncoprotein (OP). PKCa has been implicated in tumorigenesis, as it is strongly activated by many different tumour promoting compounds, such as phorbol esters and teleocidins. While these compounds promote binding of PKC isoforms to the plasma membrane of cells, which stimulates their phosphotransferase activity, these agents also induce rapid down-regulation of PKC isoforms. Consequently, it is controversial whether PKCa is an oncoprotein or a tumour suppressor protein. PKCa is highly expressed in several cancers, including malignant breast cancers and gliomas. An N294G point mutation in PKCa has been linked to the invasive phenotype of these tumours, indicating a gain-of-function in the kinase catalytic phosphotransfersase activity. Increased expression of PKCa has been observed in adenomatous pituitaries, with highest expression in invasive pituitary tumours. A point mutation was also identified in 4 invasive pituitary tumours. Furthermore, PKCa appears to stimulate cell proliferation, because many of its direct targets, such as Raf1, BCL2, and CSPG4, participate in growth factor signalling pathways and PKC activators induce the downstream activation of ERK1/2 (MAPK3/1) and RAP1GAP.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +94, p<0.057); Brain oligodendrogliomas (%CFC= -97, p<0.02); and Oral squamous cell carcinomas (OSCC) (%CFC= +57, p<0.079). The COSMIC website notes an up-regulated expression score for PKCa in diverse human cancers of 490, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 22 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25491 diverse cancer specimens. This rate is only 18 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.48 % in 805 skin cancers tested; 0.45 % in 1093 large intestine cancers tested; 0.32 % in 602 endometrium cancers tested; 0.25 % in 589 stomach cancers tested; 0.12 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,654 cancer specimens
Comments:
Only 6 deletions, no insertions or complex mutations are noted on the COSMIC website. About 25% of the point mutations are silent and do not change the amino acid sequence of the protein kinase.

