Nomenclature
Short Name:
PKD2
Full Name:
Protein kinase C, D2 type
Alias:
- DKFZP586E0820
- KPCO
- NPKC-D2
- PRKD2
- Protein kinase C, D2 type
- Protein kinase D2 homolog
- EC 2.7.11.13
- HSPC187
- Kinase PKD2
- KPCD2
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
PKD
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
96750
# Amino Acids:
878
# mRNA Isoforms:
2
mRNA Isoforms:
96,750 Da (878 AA; Q9BZL6); 80,143 Da (721 AA; Q9BZL6-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
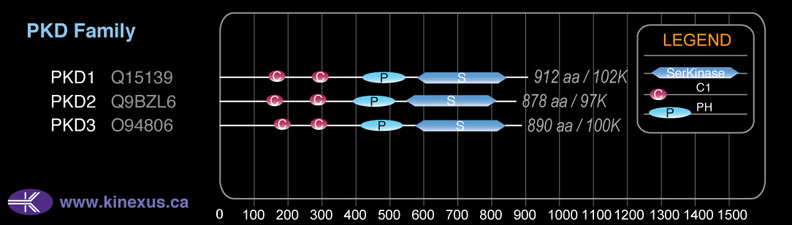
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K572, K580, K584.
Serine phosphorylated:
S189, S197, S198, S200, S203, S206, S212, S214, S225, S236, S237, S238, S239, S241, S242, S244, S245, S355, S362, S374, S375, S387, S395, S396, S518, S529, S706+, S710+, S876+.
Threonine phosphorylated:
T199, T227, T391, T392, T398, T412, T619, T714-.
Tyrosine phosphorylated:
Y246, Y272, Y378, Y438, Y717-.
Ubiquitinated:
K80, K92, K178, K286, K424, K705.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 77
77
959
38
816
 11
11
139
16
126
 13
13
160
2
126
 36
36
453
104
1022
 90
90
1121
34
798
 15
15
188
91
206
 23
23
284
41
539
 85
85
1065
31
1571
 59
59
737
17
521
 17
17
216
97
295
 14
14
169
21
160
 74
74
919
159
682
 26
26
321
24
142
 14
14
172
12
143
 21
21
258
15
318
 10
10
120
20
96
 20
20
249
113
100
 17
17
217
11
205
 8
8
105
95
87
 67
67
829
137
695
 12
12
145
13
129
 46
46
577
17
491
 14
14
170
4
109
 12
12
144
11
146
 37
37
460
13
332
 91
91
1135
63
1356
 26
26
322
27
150
 17
17
212
11
152
 17
17
209
11
137
 16
16
194
42
126
 97
97
1211
24
855
 100
100
1246
41
1853
 21
21
256
83
314
 72
72
893
78
734
 26
26
328
48
393
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 82.4
82.4
84.8
96 67.1
67.1
78.8
- -
-
-
96 -
-
-
- 96
96
97.1
96 -
-
-
- 95
95
97.1
96 94.9
94.9
97
95.5 -
-
-
- 63.8
63.8
75.3
- -
-
-
- -
-
-
- 70
70
80.5
- -
-
-
- -
-
-
- -
-
-
- 42
42
53.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20.3
20.3
34.4
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | HDAC7 - Q8WUI4 |
| 2 | TGOLN2 - O43493 |
| 3 | YWHAH - Q04917 |
| 4 | CSNK1E - P49674 |
| 5 | CSNK1D - P48730 |
| 6 | PDLIM7 - Q9NR12 |
| 7 | PTPN3 - P26045 |
| 8 | RXRA - P19793 |
| 9 | GLRX3 - O76003 |
| 10 | GRIP1 - Q9Y3R0 |
| 11 | RARB - P10826 |
| 12 | SNRNP70 - P08621 |
| 13 | HEY1 - Q9Y5J3 |
| 14 | DST - Q8WXK8 |
| 15 | DST - O94833 |
Regulation
Activation:
Activated by DAG and phorbol esters. Phorbol-ester/DAG-type domains bind DAG, mediating translocation to membranes. Autophosphorylation of Ser-710 and phosphorylation of Ser-706 by PKC relieves auto-inhibition by the PH domain
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCa | P17252 | S706 | ARIIGEKSFRRSVVG | + |
| PKCh | P24723 | S706 | ARIIGEKSFRRSVVG | + |
| PKCe | Q02156 | S706 | ARIIGEKSFRRSVVG | + |
| PKCa | P17252 | S710 | GEKSFRRSVVGTPAY | + |
| PKCh | P24723 | S710 | GEKSFRRSVVGTPAY | + |
| PKCe | Q02156 | S710 | GEKSFRRSVVGTPAY | + |
| PKCa | P17252 | S876 | QGLAERISVL_____ | + |
| PKD2 | Q9BZL6 | S876 | QGLAERISVL_____ | + |
| PKCh | P24723 | S876 | QGLAERISVL_____ | + |
| PKCe | Q02156 | S876 | QGLAERISVL_____ | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
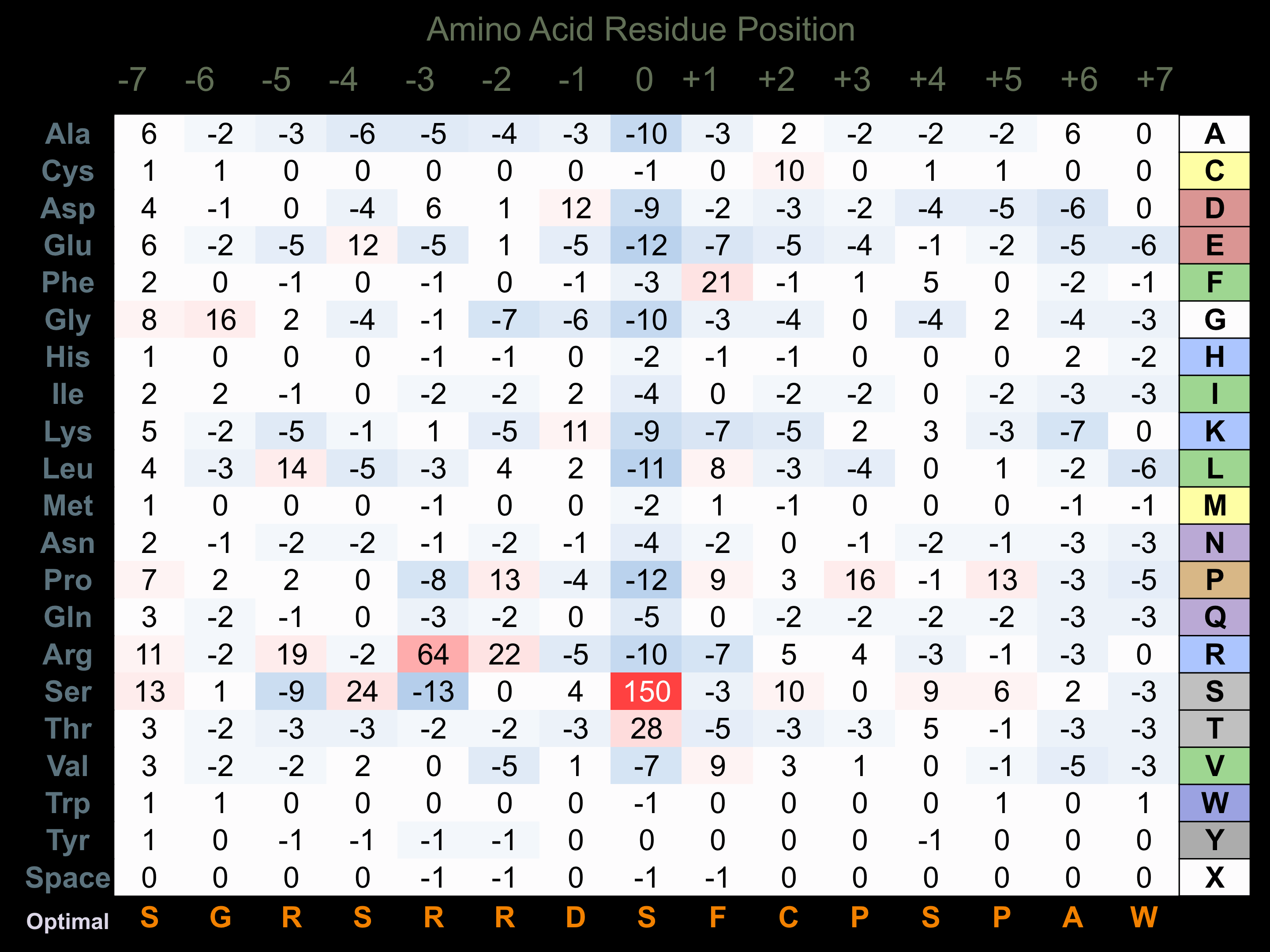
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Nephrological, neuronal, and liver disorders
Specific Diseases (Non-cancerous):
Polycystic kidney disease, Type 2 (PKD2); kidney disease; Polycystic kidney disease (PKD, PKD1); Polycystic kidney disease, autosomal dominant (ADPKD); Polycystic Liver disease (PCLD); Cystic kidney; Liver disease; Intracranial aneurysm; Polycystic kidney disease, Type 1
Comments:
Polycystic Kidney Disease, Type 2 (PKD2) is a rare nephrological disorder related to polycystic kidney disease (PKD) which is characterized by formation of cysts in the kidney, and can possibly lead to kidney failure. Kidney Disease is related to PKD2. Polycystic Kidney Disease, Autosomal Dominant (ADPKD) is a rare inherited disease characterized by development of cysts in the kidney. The rare inherited liver condition Polycystic Liver Disease (PCLD) is characterized by abdominal pain, liver swelling, and cyst development in the liver. Liver disease is also referred to as liver disorder in pregnancy and it is a rare condition that has a relation to hepatitis and fatty liver disease. Intracranial Aneurysm is characterized by the ballooning, or bulging of an artery in the brain leading to a brain aneurysm. For PKD2 the S244E, S706E, and S710E mutations, in conjunction, will lead to constitutive kinase phosphotransferase activity. A marginal increase in the ability of PKD2 to bind DAG and a great increase in PKD2 to bind phorbol ester occurs with a P275G mutation. Phosphorylation of PKD2 is lost with a Y438F mutation. Full loss of kinase phosphotransferase activity in PKD2 can occur with a D695A mutation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +61, p<0.026); Brain glioblastomas (%CFC= -69, p<0.006); Classical Hodgkin lymphomas (%CFC= -49, p<0.101); Clear cell renal cell carcinomas (cRCC) (%CFC= +52, p<0.092); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -70, p<0.0001); Large B-cell lymphomas (%CFC= +88, p<0.048); and Ovary adenocarcinomas (%CFC= +56, p<0.098). The COSMIC website notes an up-regulated expression score for PKD2 in diverse human cancers of 421, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 161 for this protein kinase in human cancers was 2.7-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24726 diverse cancer specimens. This rate is only -2 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.3 % in 1270 large intestine cancers tested; 0.28 % in 864 skin cancers tested; 0.25 % in 589 stomach cancers tested; 0.23 % in 603 endometrium cancers tested; 0.18 % in 127 biliary tract cancers tested; 0.15 % in 833 ovary cancers tested; 0.15 % in 548 urinary tract cancers tested; 0.11 % in 1634 lung cancers tested; 0.08 % in 273 cervix cancers tested; 0.05 % in 238 bone cancers tested; 0.05 % in 1512 liver cancers tested; 0.05 % in 1316 breast cancers tested; 0.04 % in 2009 haematopoietic and lymphoid cancers tested; 0.04 % in 1276 kidney cancers tested; 0.03 % in 881 prostate cancers tested; 0.03 % in 710 oesophagus cancers tested; 0.03 % in 441 autonomic ganglia cancers tested; 0.03 % in 2103 central nervous system cancers tested; 0.02 % in 558 thyroid cancers tested; 0.02 % in 1459 pancreas cancers tested.
Frequency of Mutated Sites:
None > 4 in 19,781 cancer specimens
Comments:
Only 3 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

