Nomenclature
Short Name:
PKCe
Full Name:
Protein kinase C, epsilon type
Alias:
- EC 2.7.11.13
- PKCEA
- PKC-epsilon
- PRKC
- KPCE
- MGC125656
- MGC125657
- NPKC-epsilon
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKC
SubFamily:
Eta
Specific Links
Structure
Mol. Mass (Da):
83,674
# Amino Acids:
737
# mRNA Isoforms:
1
mRNA Isoforms:
83,674 Da (737 AA; Q02156)
4D Structure:
Forms a ternary complex with TRIM63 and GN2BL1 By similarity. Interacts with DGKQ.
1D Structure:
Subfamily Alignment
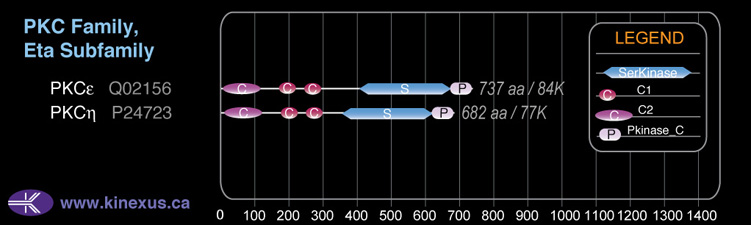
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K421, K430.
Serine phosphorylated:
S234, S261, S316, S329, S334, S337, S339, S344, S346, S350, S368, S381, S388, S729+.
Threonine phosphorylated:
T181, T228, T286, T309+, T563+, T564+, T565+, T566+, T570-, T710.
Tyrosine phosphorylated:
Y434, Y573-.
Ubiquitinated:
K321.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1181
31
1045
 5
5
58
12
77
 5
5
56
14
55
 23
23
266
155
528
 44
44
515
38
360
 1.4
1.4
16
64
13
 14
14
167
53
369
 31
31
365
52
911
 16
16
191
10
167
 11
11
126
151
192
 5
5
55
37
74
 41
41
486
157
598
 7
7
77
25
77
 4
4
50
8
53
 7
7
85
21
136
 1.4
1.4
16
22
16
 1.3
1.3
15
414
21
 4
4
52
20
94
 4
4
51
116
51
 35
35
419
140
378
 5
5
57
35
63
 4
4
48
38
62
 4
4
45
23
44
 5
5
57
20
53
 4
4
45
33
70
 54
54
632
96
1089
 5
5
56
28
59
 4
4
53
21
50
 5
5
59
19
43
 4
4
43
56
32
 58
58
690
18
483
 26
26
302
42
435
 35
35
409
90
1039
 57
57
679
109
645
 20
20
237
61
465
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 62.3
62.3
75.8
100 99.7
99.7
99.9
100 -
-
-
99 -
-
-
- 98.9
98.9
99.3
99 -
-
-
- 98
98
99.5
98 98.1
98.1
99.3
98 -
-
-
- -
-
-
- 94.6
94.6
96.8
96 -
-
-
92 36.1
36.1
51.4
87 -
-
-
- 52.8
52.8
66.2
60 -
-
-
- 58.6
58.6
74.8
62 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
41 -
-
-
44.5
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PRKD1 - Q15139 |
| 2 | ACTA1 - P68133 |
| 3 | COPB2 - P35606 |
| 4 | RAF1 - P04049 |
| 5 | PIK3CB - P42338 |
| 6 | SLC4A3 - P48751 |
| 7 | ITGB1 - P05556 |
| 8 | CNN1 - P51911 |
| 9 | PDPK1 - O15530 |
| 10 | TRPV1 - Q8NER1 |
| 11 | SRC - P12931 |
| 12 | ACTC1 - P68032 |
| 13 | MAPK1 - P28482 |
| 14 | BAX - Q07812 |
| 15 | PDPK2 - Q6A1A2 |
Regulation
Activation:
Phosphorylation at Thr-566 increases phosphotransferase activity and autophosphorylation at Ser-279.Phosphorylation of Ser-729 increases phosphotransferase activity, promotes dephosphorylation of Thr-566 and possibly nuclear import. Thr-566 (activation loop of the kinase domain), Thr-710 (turn motif) and Ser-729 (hydrophobic region), need to be phosphorylated for its full activation.
Inhibition:
Phosphorylation at Ser-368 induces interaction with 14-3-3 beta.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PKCe | Q02156 | S234 | ETPDQVGSQRFSVNM | |
| PKCa | P17252 | S234 | ETPDQVGSQRFSVNM | |
| PKCe | Q02156 | S316 | TPDKITNSGQRRKKL | |
| PKCa | P17252 | S316 | TPDKITNSGQRRKKL | |
| GSK3B | P49841 | S346 | SEEDRSKSAPTSPCD | ? |
| p38a | Q16539 | S350 | RSKSAPTSPCDQEIK | |
| p38b | Q15759 | S350 | RSKSAPTSPCDQEIK | |
| PKCe | Q02156 | S368 | NNIRKALSFDNRGEE | ? |
| PKCa | P17252 | S368 | NNIRKALSFDNRGEE | ? |
| PDK1 | O15530 | T566 | LNGVTTTTFCGTPDY | + |
| PKCe | Q02156 | S729 | QEEFKGFSYFGEDLM | + |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ADAM17 | P78536 | T735 | KPFPAPQTPGRLQPA | |
| Akt1 (PKBa) | P31749 | S473 | RPHFPQFSYSASGTA | + |
| Akt1 (PKBa) | P31749 | T308 | KDGATMKTFCGTPEY | + |
| CACNA1B | Q00975 | S1757 | FEMLKHMSPPLGLGK | |
| CACNA1B | Q00975 | S2111 | RRQERGRSQERRQPS | |
| CACNA1B | Q00975 | S424 | KRAATKKSRNDLIHA | |
| CACNA1B | Q00975 | T421 | DVLKRAATKKSRNDL | |
| CACNA1D | Q01668 | S81 | PPPVGSLSQRKRQQY | |
| ChAT | P28329 | S558 | VPTYESASIRRFQEG | |
| ChAT | P28329 | S594 | HKAAVPASEKLLLLK | |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| Cx43 | P17302 | S363 | AIVDQRPSSRASSRA | |
| Cx43 | P17302 | S364 | IVDQRPSSRASSRAS | |
| Cx43 | P17302 | S367 | QRPSSRASSRASSRP | |
| Cx43 | P17302 | S368 | RPSSRASSRASSRPR | |
| Cx43 | P17302 | S371 | SRASSRASSRPRPDD | |
| Cx43 | P17302 | S372 | RASSRASSRPRPDDL | |
| GAP43 | P17677 | S41 | AATKIQASFRGHITR | |
| Gg(12) | Q9UBI6 | S2 | _______SSKTASTN | |
| IQGAP1 | P46940 | S1443 | DKMKKSKSVKEDSNL | |
| IRS1 | P35568 | S323 | MVGGKPGSFRVRASS | |
| ITGB1 | P05556 | T788 | PIYKSAVTTVVNPKY | |
| ITGB1 | P05556 | T789 | IYKSAVTTVVNPKYE | |
| KIR3DL1 | P43629 | S415 | QRKITRPSQRPKTPP | |
| KRT18 | P05783 | S53 | ISVSRSTSFRGGMGS | |
| KRT8 | P05787 | S24 | PRAFSSRSYTSGPGS | |
| KRT8 | P05787 | S9 | SIRVTQKSYKVSTSG | |
| Met | P08581 | S985 | PHLDRLVSARSVSPT | - |
| MYBPC3 | Q14896 | S304 | SLLKKSSSFRTPRDS | |
| Occludin | Q16625 | T403 | DHYETDYTTGGESCD | |
| Occludin | Q16625 | T404 | HYETDYTTGGESCDE | |
| p70S6Kb (RPS6KB2) | Q9UBS0 | S473 | PPSGTKKSKRGRGRP | + |
| PKCd (PRKCD) | Q05655 | T507 | FGESRASTFCGTPDY | + |
| PKCe (PRKCE) | Q02156 | S234 | ETPDQVGSQRFSVNM | |
| PKCe (PRKCE) | Q02156 | S316 | TPDKITNSGQRRKKL | |
| PKCe (PRKCE) | Q02156 | S368 | NNIRKALSFDNRGEE | ? |
| PKCe (PRKCE) | Q02156 | S729 | QEEFKGFSYFGEDLM | + |
| PKD1 (PRKCM) | Q15139 | S738 | ARIIGEKSFRRSVVG | + |
| PKD1 (PRKCM) | Q15139 | S742 | GEKSFRRSVVGTPAY | + |
| PKD2 (PRKD2) | Q9BZL6 | S706 | ARIIGEKSFRRSVVG | + |
| PKD2 (PRKD2) | Q9BZL6 | S710 | GEKSFRRSVVGTPAY | + |
| PKD2 (PRKD2) | Q9BZL6 | S876 | QGLAERISVL_____ | + |
| PLCB3 | Q01970 | S1105 | LDRKRHNSISEAKMR | - |
| PLM | O00168 | S83 | EEGTFRSSIRRLSTR | |
| PLM | O00168 | S88 | RSSIRRLSTRRR___ | |
| PLM | O00168 | T89 | SSIRRLSTRRR____ | |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| RIP140 | P48552 | S1001 | CMDNRTFSYPGVVKT | |
| RIP140 | P48552 | S102 | AAKRKRLSDSIMNLN | |
| SLC24A2 | Q9UI40 | S495 | PDVRKPSSRKFFPIT | |
| SLC24A2 | Q9UI40 | T165 | VPSLTVITEKLGISD | |
| SLC24A2 | Q9UI40 | T467 | SLAWPSETRKQVTFL | |
| SLC6A2 | P23975 | S259 | LWKGVKTSGKVVWIT | |
| STAT3 | P40763 | S727 | NTIDLPMSPRALDSL | - |
| Tau iso8 | P10636-8 | S258 | PDLKNVKSKIGSTEN | |
| Tau iso8 | P10636-8 | S293 | NVQSKCGSKDNIKHV | |
| Tau iso8 | P10636-8 | S324 | KVTSKCGSLGNIHHK | |
| Tau iso8 | P10636-8 | S352 | DFKDRVQSKIGSLDN | |
| TICAM-2 | Q86XR7 | S16 | NSCPLSLSWGKRHSV | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TNNI3 | P19429 | S41 | AKKKSKISASRKLQL | |
| TNNI3 | P19429 | S43 | KKSKISASRKLQLKT | |
| TNNI3 | P19429 | S76 | GEKGRALSTRCQPLE | |
| TNNI3 | P19429 | T142 | RGKFKRPTLRRVRIS | |
| TNNT2 | P45379 | S207 | QAQTERKSGKRQTER | |
| TNNT2 | P45379 | T203 | YIQKQAQTERKSGKR | |
| TNNT2 | P45379 | T212 | RKSGKRQTEREKKKK | |
| TNNT2 | P45379 | T293 | TRGKAKVTGRWK___ | |
| UGT1A7 | Q9HAW7 | S432 | KAVINDKSYKENIMR | |
| VR1 | Q8NER1 | S502 | YFLQRRPSMKTLFVD | |
| VR1 | Q8NER1 | S801 | VPLLREASARDRQSA |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
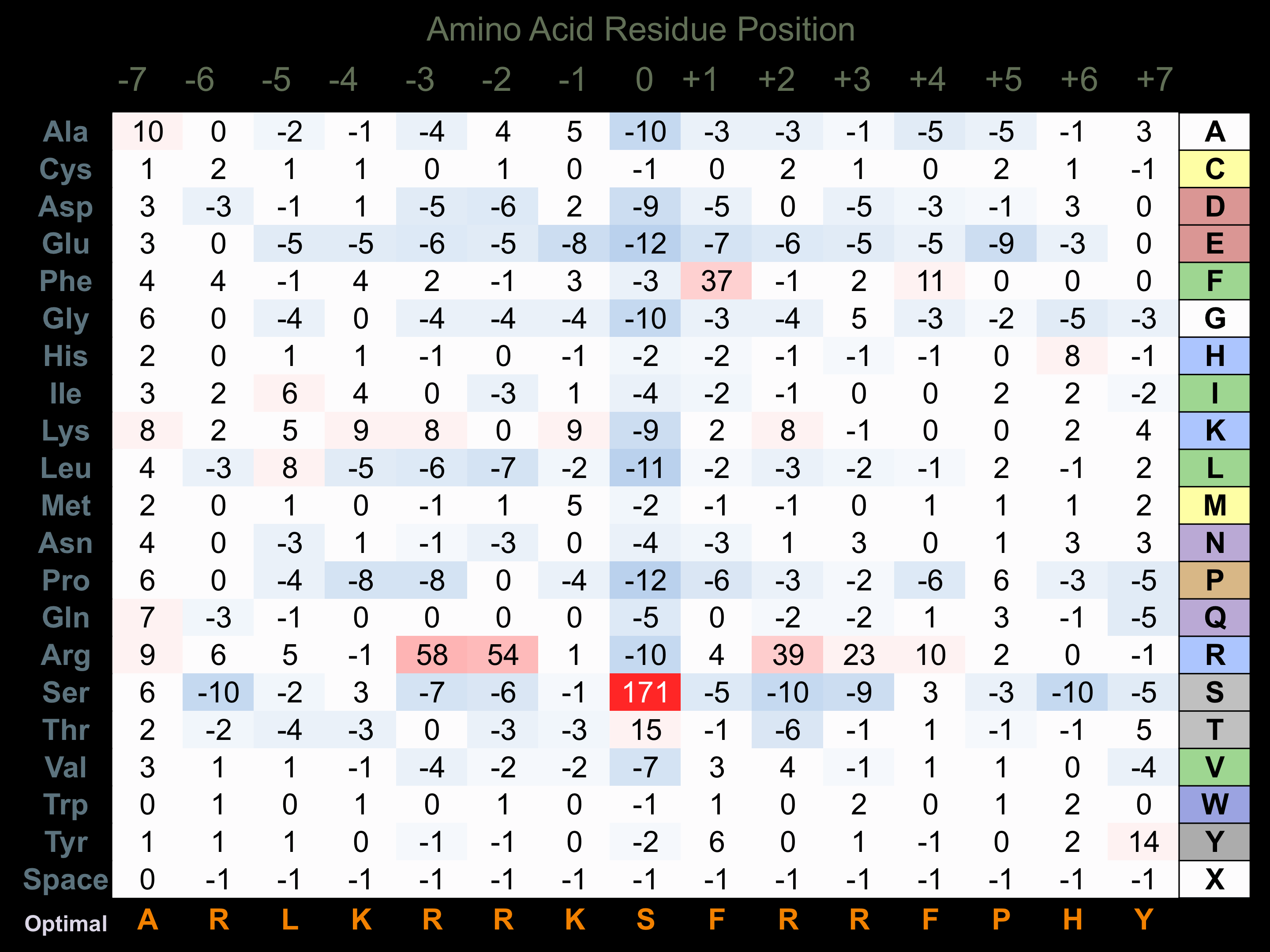
Matrix Type:
Experimentally derived from alignment of 121 known protein substrate phosphosites and 50 peptides phosphorylated by recombinant PKCe in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cardiovascular disorders
Specific Diseases (Non-cancerous):
Ischemia; Rift Valley fever
Comments:
Activation of PKCe has been demonstrated to be cardioprotective in animal models of myocardial ischemia and reperfusion injury, potentially suggesting a role for the PKCe protein in cellular anti-oxidant mechanisms. In addition, PKCe is hypothesized to interact with components of the mitochondrial transition pore, specifically Vdac, and mediate inhibition of Ca2+ induced pore opening. Loss-of-function mutations in this protein are linked with ischemia, presumably through the loss of the protective anti-oxidant function of the protein, leading to exaggerated oxidative tissue damage. Furthermore, mutations in the PKCe gene are linked to Rift Valley Fever, a viral infection transmitted by aedes mosquitoes, which has symptoms including vomiting blood, jaundice, bleeding from the nose or gums, menorrhagia, and bleeding from venepuncture sites.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -47, p<0.013); Bladder carcinomas (%CFC= -46, p<0.0002); and Brain oligodendrogliomas (%CFC= -96, p<0.068). The COSMIC website notes an up-regulated expression score for PKCe in diverse human cancers of 462, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 17 for this protein kinase in human cancers was 0.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25409 diverse cancer specimens. This rate is only -8 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.39 % in 589 stomach cancers tested; 0.35 % in 1296 large intestine cancers tested; 0.25 % in 603 endometrium cancers tested; 0.13 % in 710 oesophagus cancers tested; 0.12 % in 1276 kidney cancers tested; 0.1 % in 1316 breast cancers tested; 0.09 % in 891 ovary cancers tested; 0.09 % in 864 skin cancers tested; 0.09 % in 1956 lung cancers tested; 0.06 % in 942 upper aerodigestive tract cancers tested; 0.06 % in 238 bone cancers tested; 0.05 % in 273 cervix cancers tested; 0.04 % in 382 soft tissue cancers tested; 0.02 % in 558 thyroid cancers tested; 0.02 % in 548 urinary tract cancers tested; 0.02 % in 2009 haematopoietic and lymphoid cancers tested; 0.01 % in 2103 central nervous system cancers tested; 0.01 % in 1512 liver cancers tested; 0.01 % in 1467 pancreas cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R502* (3).
Comments:
Only 3 deletions, no insertions, and no mutations are noted on the COSMIC website.

