Nomenclature
Short Name:
PKCz
Full Name:
Protein kinase C, zeta type
Alias:
- EC 2.7.11.13
- PKC-zeta
- PKMzeta
- PRKCZ
- Protein kinase C zeta
- Protein kinase M zeta
- Kinase PKC-zeta
- KPCZ
- NPKC-zeta
- PKC2
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKC
SubFamily:
Iota
Specific Links
Structure
Mol. Mass (Da):
67,660
# Amino Acids:
592
# mRNA Isoforms:
3
mRNA Isoforms:
67,660 Da (592 AA; Q05513); 56,136 Da (488 AA; Q05513-3); 46,622 Da (409 AA; Q05513-2)
4D Structure:
Forms a ternary complex with SQSTM1 and KCNAB2. Forms another ternary complex with SQSTM1 and GABRR3. Forms a complex with SQSTM1 and MAP2K5 By similarity. Interacts with PARD6A, PARD6B, PARD6G and SQSTM1. Part of a complex with PARD3, PARD6A or PARD6B or PARD6G and CDC42 or RAC1. Interacts with ADAP1/CENTA1. Forms a ternary complex composed of SQSTM1 and PAWR. Interacts directly with SQSTM1 Probable. Interacts with IKBKB. Interacts (via the protein kinase domain) with WWC1. Forms a tripartite complex with WWC1 and DDR1, but predominantly in the absence of collagen
1D Structure:
Subfamily Alignment
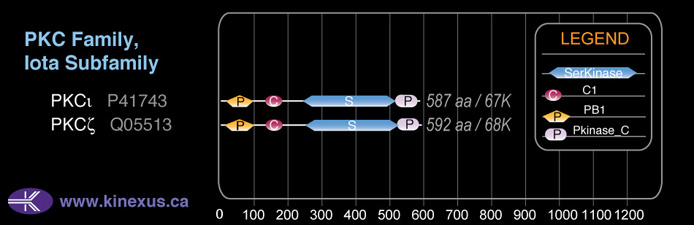
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S113, S186, S190, S223, S228, S262, S409+.
Threonine phosphorylated:
T407+, T408+, T410+, T414-, T560-.
Tyrosine phosphorylated:
Y263-, Y356+, Y417-, Y428.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 51
51
577
25
987
 7
7
82
11
60
 1.3
1.3
15
1
0
 68
68
772
78
840
 59
59
668
21
560
 11
11
127
50
184
 23
23
261
29
372
 94
94
1063
22
1954
 73
73
822
10
668
 18
18
205
47
103
 6
6
63
16
54
 73
73
825
103
717
 7
7
84
12
17
 1.4
1.4
16
7
10
 4
4
42
13
21
 9
9
102
13
99
 18
18
207
175
1582
 5
5
52
8
29
 0.7
0.7
8
43
7
 52
52
590
79
540
 26
26
289
12
242
 10
10
118
14
130
 7
7
74
10
32
 39
39
443
8
269
 4
4
47
12
17
 78
78
885
43
1126
 9
9
105
15
36
 7
7
81
8
29
 2
2
24
8
22
 5
5
51
14
28
 63
63
713
42
431
 43
43
482
26
579
 14
14
163
91
597
 60
60
678
57
640
 100
100
1132
35
1611
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 91.4
91.4
93.1
100 70
70
81.5
- -
-
-
93 -
-
-
99 94.7
94.7
97.1
95 -
-
-
- 95.4
95.4
96.9
96 95.9
95.9
97.3
96 -
-
-
- -
-
-
- 87.9
87.9
92.7
90 33.1
33.1
50.1
86 70.7
70.7
81.9
82 -
-
-
- 36.5
36.5
54.8
- -
-
-
- 56.6
56.6
70.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PARD6A - Q9NPB6 |
| 2 | AKT3 - Q9Y243 |
| 3 | RELA - Q04206 |
| 4 | YWHAZ - P63104 |
| 5 | STAT3 - P40763 |
| 6 | PPP3CA - Q08209 |
| 7 | DDX17 - Q92841 |
| 8 | VHL - P40337 |
| 9 | FEZ1 - Q99689 |
| 10 | KRT10 - P13645 |
| 11 | GSK3B - P49841 |
| 12 | IRS1 - P35568 |
| 13 | PAWR - Q96IZ0 |
| 14 | GSK3A - P49840 |
| 15 | PSEN1 - P49768 |
Regulation
Activation:
Phosphatidylinositol 3,4,5-trisphosphate might be a physiological activator. Phosphorylation at Thr-410 (activation loop of the kinase domain) and Thr-560 (turn motif) increases phosphotransferase activity. Phosphorylation of Tyr-356 induces interaction with 14-3-3 to increase PKCz phosphotransferase activity and facilitate its complex formation with Raf1 to activate it. Phosphorylation of Ser-113, Ser-148, Thr-179, Ser-186, Ser-217, Ser-218, Ser-262, Ser-520 induces interaction with 14-3-3 beta and Raf1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ADD1 | P35611 | S726 | KKKFRTPSFLKKSKK | |
| ADD2 | P35612 | S713 | KKKFRTPSFLKKSKK | |
| Akt1 (PKBa) | P31749 | T34 | FLLKNDGTFIGYKER | - |
| AQP9 | O43315 | S11 | EGAEKGKSFKQRLVL | |
| AQP9 | O43315 | S222 | MNPARDLSPRLFTAL | |
| ATP1A1 | P05023 | S16 | KYEPAAVSEQGDKKG | |
| Baxa | Q07812 | S184 | VAGVLTASLTIWKKM | - |
| CABP4 | P57796 | S42 | PPLTRKRSKKERGLR | |
| Caspase 9 | P55211 | S144 | GDVGALESLRGNADL | |
| ChAT | P28329 | S558 | VPTYESASIRRFQEG | |
| ChAT | P28329 | S594 | HKAAVPASEKLLLLK | |
| Gem | P55040 | S261 | AYQKRKESMPRKARR | |
| GSK3a | P49840 | S21 | SGRARTSSFAEPGGG | - |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| IKKb (IKBKINASE) | O14920 | S177 | AKELDQGSLCTSFVG | + |
| IKKb (IKBKINASE) | O14920 | S181 | DQGSLCTSFVGTLQY | + |
| IRAP | Q9UIQ6 | S80 | SAKLLGMSFMNRSSG | |
| IRAP | Q9UIQ6 | S91 | RSSGLRNSATGYRQS | |
| IRS1 | P35568 | S24 | GYLRKPKSMHKRFFV | |
| IRS1 | P35568 | S312 | TESITATSPASMVGG | - |
| IRS1 | P35568 | S323 | MVGGKPGSFRVRASS | |
| IRS1 | P35568 | S441 | SPCDFRSSFRSVTPD | |
| IRS1 | P35568 | S503 | PGTGLGTSPALAGDE | |
| IRS1 | P35568 | S574 | RLPGHRHSAFVPTRS | |
| IRS1 | P35568 | S616 | DDGYMPMSPGVAPVP | - |
| LKB1 (STK11) | Q15831 | S307 | IRQIRQHSWFRKKHP | ? |
| LKB1 (STK11) | Q15831 | S428 | SSKIRRLSACKQQ__ | + |
| MARK1 | Q9P0L2 | T613 | RGSSSRSTFHGEQLR | - |
| MARK2 | Q7KZI7 | T596 | RGVSSRSTFHAGQLR | - |
| MARK3 | P27448 | T564 | RGTASRSTFHGQPRE | |
| MARK3 | P27448 | T587 | RGTASRSTFHGQPRE | |
| Met | P08581 | S985 | PHLDRLVSARSVSPT | - |
| MYH10 | P35580 | S1937 | RGGPISFSSSRSGRR | |
| NCOA3 (SRC-3) | Q9Y6Q9 | S1033 | NRPLLRNSLDDLVGP | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S1042 | DDLVGPPSNLEGQSD | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S1048 | PSNLEGQSDERALLD | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | S1062 | DQLHTLLSNTDATGL | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | T1059 | ALLDQLHTLLSNTDA | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | T1064 | LHTLLSNTDATGLEE | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | T1067 | LLSNTDATGLEEIDR | - |
| NCOA3 (SRC-3) | Q9Y6Q9 | T1114 | KAGLYGQTYPAQGPP | - |
| NFkB-p65 | Q04206 | S311 | RTYETFKSIMKKSPF | |
| NR1H4 iso2 | Q96RI1-2 | T442 | GRLTELRTFNHHHAE | |
| p47phox | P14598 | S303 | RGAPPRRSSIRNAHS | + |
| p47phox | P14598 | S304 | GAPPRRSSIRNAHSI | + |
| p47phox | P14598 | S315 | AHSIHQRSRKRLSQD | ? |
| p70S6Kb (RPS6KB2) | Q9UBS0 | S473 | PPSGTKKSKRGRGRP | + |
| PARD3 | Q8TEW0 | S827 | REGFGRQSMSEKRTK | |
| PFKFB2 | O60825 | T475 | TPLSSSNTIRRPRNY | |
| PKCz (PRKCZ) | Q05513 | S113 | PCPGEDKSIYRRGAR | |
| PKCz (PRKCZ) | Q05513 | S186 | TCRKHMDSVMPSQEP | |
| PKCz (PRKCZ) | Q05513 | T560 | TSEPVQLTPDDEDAI | ? |
| PKD1 (PRKCM) | Q15139 | S738 | ARIIGEKSFRRSVVG | + |
| PKD1 (PRKCM) | Q15139 | S742 | GEKSFRRSVVGTPAY | + |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PTEN | P60484 | S380 | EPDHYRYSDTTDSDP | - |
| PTEN | P60484 | T382 | DHYRYSDTTDSDPEN | - |
| PTEN | P60484 | T383 | HYRYSDTTDSDPENE | - |
| SP1 | P08047 | S641 | GKVYGKTSHLRAHLR | |
| SP1 | P08047 | S670 | CGKRFTRSDELQRHK | |
| SP1 | P08047 | T668 | SYCGKRFTRSDELQR | |
| SP1 | P08047 | T681 | QRHKRTHTGEKKFAC | |
| Tau iso8 | P10636-8 | S258 | PDLKNVKSKIGSTEN | |
| Tau iso8 | P10636-8 | S293 | NVQSKCGSKDNIKHV | |
| Tau iso8 | P10636-8 | S324 | KVTSKCGSLGNIHHK | |
| Tau iso8 | P10636-8 | S352 | DFKDRVQSKIGSLDN | |
| TNNI3 | P19429 | S41 | AKKKSKISASRKLQL | |
| TNNI3 | P19429 | S43 | KKSKISASRKLQLKT | |
| TRAF2 | Q12933 | S55 | QCGHRYCSFCLASIL | |
| WWC1 | Q8IX03 | S975 | VRMKRPSSVKSLRSE | |
| WWC1 | Q8IX03 | S978 | KRPSSVKSLRSERLI |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
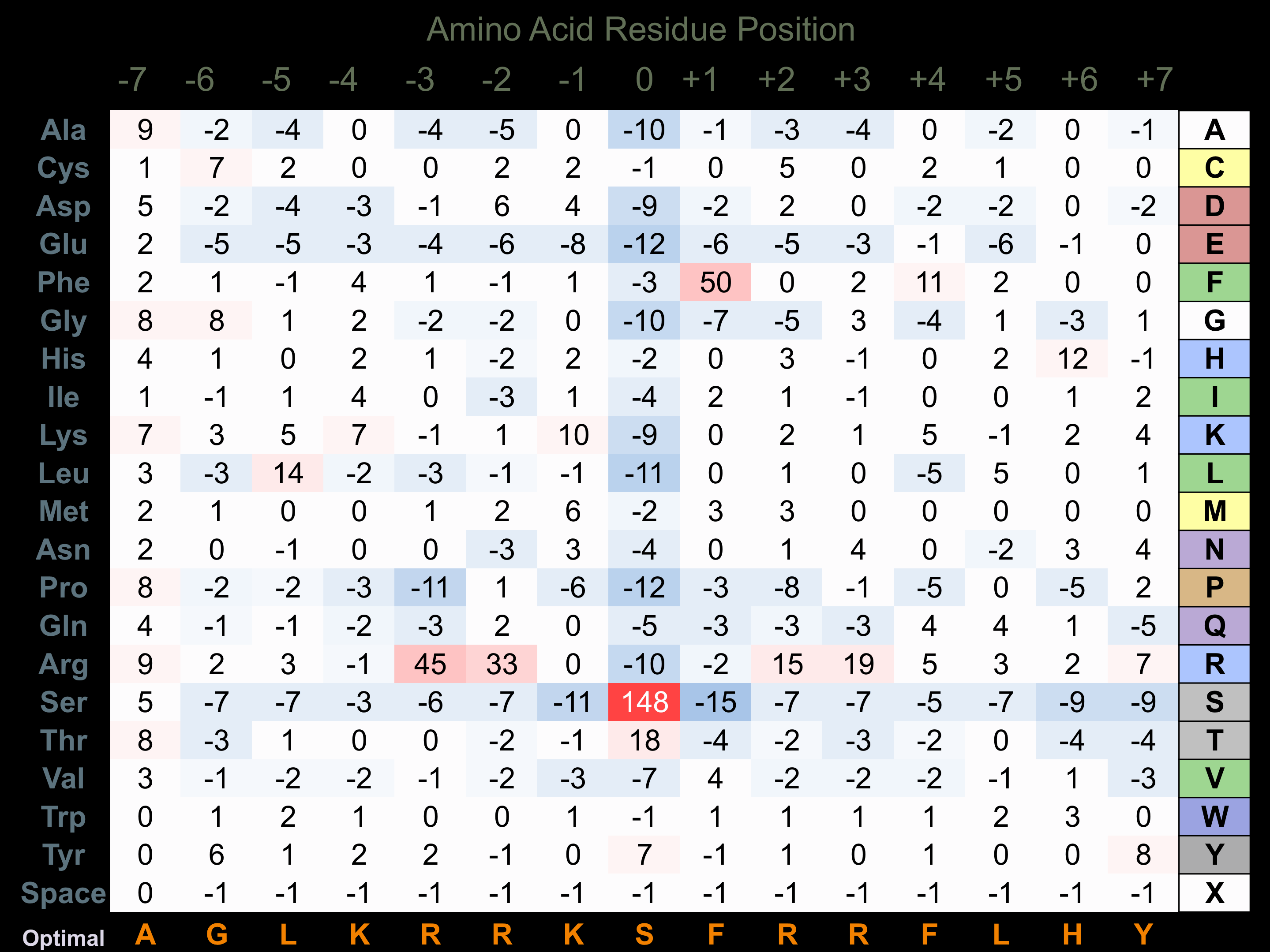
Matrix Type:
Experimentally derived from alignment of 95 known protein substrate phosphosites and 51 peptides phosphorylated by recombinant PKCz in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Infectious disease
Specific Diseases (Non-cancerous):
Parainfluenza virus Type 3 (PIV3)
Comments:
PKCz may be associated with development of allergic airway inflammation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +122, p<0.0002); Breast epithelial cell carcinomas (%CFC= +66, p<0.002); Classical Hodgkin lymphomas (%CFC= +66, p<0.066); Large B-cell lymphomas (%CFC= +74, p<0.076); Skin melanomas - malignant (%CFC= -71, p<0.0001); and T-cell prolymphocytic leukemia (%CFC= +53, p<0.002). The COSMIC website notes an up-regulated expression score for PKCz in diverse human cancers of 378, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 29 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25546 diverse cancer specimens. This rate is only -10 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.42 % in 1119 large intestine cancers tested; 0.34 % in 805 skin cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: N334S (4).
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

