Nomenclature
Short Name:
PCTAIRE3
Full Name:
Serine-threonine-protein kinase PCTAIRE-3
Alias:
- CDK18
- EC 2.7.11.22
- KPT3
- PCTAIRE protein kinase 3
- PCTK3
- Serine/threonine-protein kinase PCTAIRE-3
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
TAIRE
Specific Links
Structure
Mol. Mass (Da):
54180
# Amino Acids:
472
# mRNA Isoforms:
3
mRNA Isoforms:
57,577 Da (504 AA; Q07002-3); 54,424 Da (474 AA; Q07002-2); 54,180 Da (472 AA; Q07002)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 142 | 423 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K93.
Methylated:
R463.
Serine phosphorylated:
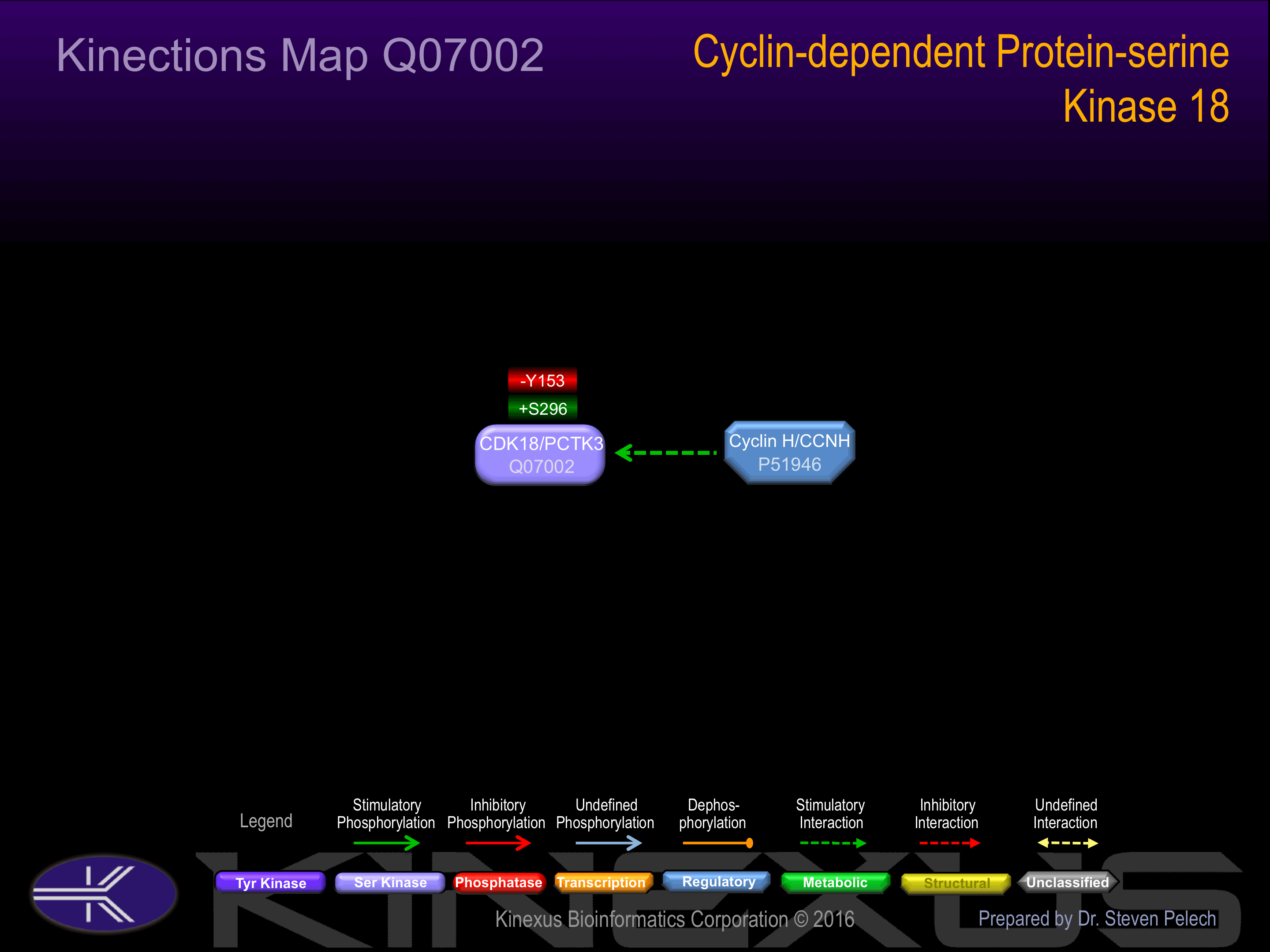
S12+, S14+, S72, S87+, S92, S96, S115, S123, S130+, S132, S296+, S470.
Threonine phosphorylated:
T141, T152, T155 .
Tyrosine phosphorylated:
Y142, Y153-.
Ubiquitinated:
K144, K171, K278.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 47
47
689
16
965
 2
2
35
10
24
 3
3
45
1
0
 29
29
417
53
459
 23
23
328
14
329
 3
3
50
41
101
 51
51
744
27
796
 42
42
608
28
696
 32
32
464
10
441
 14
14
203
70
176
 4
4
56
20
38
 27
27
393
148
586
 2
2
25
12
7
 2
2
26
10
20
 6
6
92
16
86
 5
5
67
8
16
 7
7
99
265
1048
 6
6
88
12
83
 2
2
24
55
21
 27
27
387
56
427
 32
32
463
18
366
 3
3
48
19
31
 4
4
59
10
41
 2
2
26
7
27
 2
2
34
15
25
 100
100
1456
30
3041
 2
2
25
15
12
 3
3
40
11
25
 2
2
34
9
32
 3
3
47
14
45
 25
25
364
18
257
 77
77
1122
21
1596
 15
15
212
47
416
 52
52
758
31
755
 8
8
121
22
125
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 44.3
44.3
53.1
94 61.4
61.4
75.2
97 -
-
-
92.6 -
-
-
- 61.3
61.3
73.9
90.5 -
-
-
- 87.7
87.7
92.5
91 88.1
88.1
92.8
92 -
-
-
- 63.8
63.8
77.4
- 76.4
76.4
81.9
85 46.1
46.1
61.6
79 43.8
43.8
57.4
80 -
-
-
- 34.1
34.1
44
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 33
33
47
- -
-
-
- 33.2
33.2
47.8
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | SEC23A - Q15436 |
| 2 | YWHAH - Q04917 |
| 3 | CCNB2 - O95067 |
| 4 | CCNA2-P20248 |
| 5 | CCNE1-P24864 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Tau iso5 (Tau-C) | P10636-5 | S231 | KKVAVVRTPPKSPSS | |
| Tau iso5 (Tau-C) | P10636-5 | S235 | VVRTPPKSPSSAKSR |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
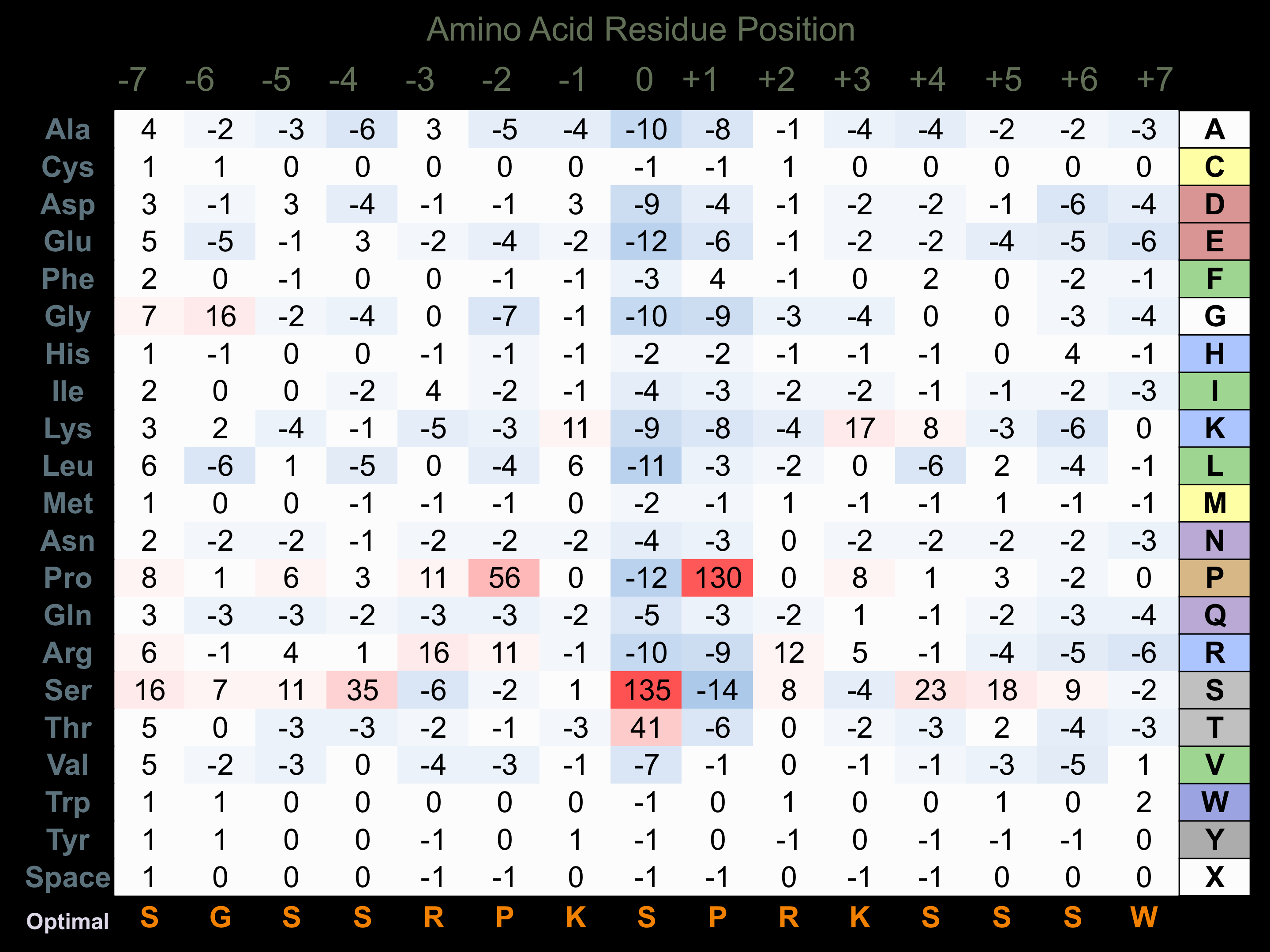
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AT7519 | Kd = 14 nM | 11338033 | 22037378 | |
| R547 | Kd = 20 nM | 6918852 | 22037378 | |
| AC1NS7CD | Kd = 44 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 44 nM | 3025986 | 296468 | 18183025 |
| AST-487 | Kd = 54 nM | 11409972 | 574738 | 18183025 |
| CHEMBL1082152 | Kd = 110 nM | 11560568 | 1082152 | 20138512 |
| Foretinib | Kd = 250 nM | 42642645 | 1230609 | 22037378 |
| Staurosporine | Kd = 270 nM | 5279 | 18183025 | |
| Nintedanib | Kd = 510 nM | 9809715 | 502835 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Alvocidib | Kd = 1.1 µM | 9910986 | 428690 | 18183025 |
| PHA-665752 | Kd = 1.5 µM | 10461815 | 450786 | 22037378 |
| Sunitinib | Kd = 1.7 µM | 5329102 | 535 | 18183025 |
| JNJ-7706621 | Kd = 2.1 µM | 5330790 | 191003 | 18183025 |
| TG101348 | Kd = 2.7 µM | 16722836 | 1287853 | 22037378 |
| SU14813 | Kd = 3.2 µM | 10138259 | 1721885 | 22037378 |
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Alzheimer's disease (AD)
Comments:
PCTAIRE3 has been implicated in the regulation of tau phosphorylation. Alzheimer's disease (AD) is a neurodegenerative disease characterized by the progressive loss of memory, judgement, and other cognitive processes. The hallmark of AD pathology is the deposition of amyloid-beta plaques and tau neurofibrillary tangles. These abnormalities are implicated in the disruption of cellular communication, oxidative cell damage, and eventual cell death. Multiple genes are thought to contribute to AD suceptibility along with epigenetic and environmental factors. Significantly elevated levels of PCTAIRE3 in the temporal cortex have been observed in post-mortem brain tissue from AD patients compared to control patients. In addition, the PCTAIRE3 protein was found to be concentrated in close proximity to the neurofibrillary tangles, indicating a direct interaction between the tau proteins and PCTAIRE3. Overexpression of PCTAIRE3 in cell cultures revealed that PCTAIRE3 is able to indirectly promote the phosphorylation of the tau protein at the Thr-231 and Ser-235 residues. These residues have been implicated in the early pathogenesis of AD, indicating that abnormal PCTAIRE3 expression may contribute to AD pathology.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +51, p<0.024); Bladder carcinomas (%CFC= +51, p<0.024); Breast epithelial carcinomas (%CFC= -79, p<0.004); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -65, p<0.033); Clear cell renal cell carcinomas (cRCC) (%CFC= +148, p<0.015); Pituitary adenomas (ACTH-secreting) (%CFC= -80). The COSMIC website notes an up-regulated expression score for PCTAIRE3 in diverse human cancers of 559, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 24 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24914 diverse cancer specimens. This rate is very similar (+ 3% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 3 in 20,197 cancer specimens
Comments:
Only 4 deletions, and no insertions or complex mutations are noted on the COSMIC website.