Nomenclature
Short Name:
TIF1b
Full Name:
Transcription intermediary factor 1-beta
Alias:
- KAP-1
- KRIP-1
- TRIM28
- RING finger protein 96
- RNF96
- TIF1-beta
- Tripartite motif-containing protein 28
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
TIF1
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
88,550
# Amino Acids:
835
# mRNA Isoforms:
2
mRNA Isoforms:
88,550 Da (835 AA; Q13263); 79,474 Da (753 AA; Q13263-2)
4D Structure:
Associated with HP1 alpha (CBX5), beta (CBX1) and gamma (CBX3) in interphase nuclei By similarity. Interacts with ZNF382; enhances ZNF382 transcriptional repressor activity By similarity. Interacts with CEBPB and NR3C1 By similarity. Interacts with NCOR1
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
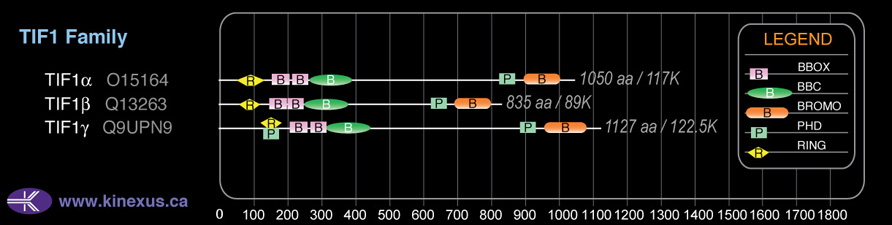
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K266 (N6), K304 (N6), K340 (N6), K377 (N6), K469, K575, K770, K774 (N6), K779.
Serine phosphorylated:
S4, S9, S14, S17, S19, S26, S37, S45, S49, S50, S138, S258, S355, S417, S437, S439, S440, S453, S459, S460, S466, S471, S473+, S479, S489, S501, S594, S596, S598, S600, S601, S612, S624, S681, S683, S689, S697, S752, S756, S757, S784, S816, S823, S824+, S828.
Sumoylated:
K554, K750, K779, K804.
Threonine phosphorylated:
T34, T498, T514, T531, T536, T541, T599, T611, T620, T690.
Tyrosine phosphorylated:
Y133, Y242, Y369, Y449+, Y458+, Y517+, Y755.
Ubiquitinated:
K127, K213, K238, K254, K261, K266, K272, K275, K289, K296, K304, K308, K319, K337, K340, K365, K366, K377, K390, K400, K407, K434, K469, K507, K575, K750, K770, K774, K779, K804.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 45
45
1149
16
1089
 12
12
309
10
206
 21
21
536
1
0
 41
41
1049
46
1202
 46
46
1158
14
989
 11
11
290
42
136
 21
21
533
19
641
 100
100
2543
21
4043
 33
33
837
10
716
 14
14
348
45
188
 12
12
301
14
193
 41
41
1041
105
593
 16
16
414
12
112
 19
19
481
9
457
 13
13
340
11
268
 22
22
558
8
180
 24
24
623
100
207
 10
10
256
8
264
 15
15
379
64
217
 37
37
943
56
792
 14
14
365
10
315
 27
27
679
12
649
 25
25
631
10
212
 28
28
723
8
517
 29
29
740
10
656
 75
75
1903
27
2975
 18
18
465
15
264
 11
11
272
8
272
 23
23
579
8
564
 0.9
0.9
23
14
13
 49
49
1257
18
498
 23
23
587
21
670
 71
71
1796
54
1848
 38
38
954
31
517
 6
6
140
22
90
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 25.1
25.1
37
100 88.1
88.1
88.4
- -
-
-
96 -
-
-
98 95.9
95.9
97
97 -
-
-
- 94.1
94.1
96.9
94 92.3
92.3
95.8
93.5 -
-
-
- 31.4
31.4
46.3
- -
-
-
- 29.1
29.1
44
59.5 27
27
40.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CBX5 - P45973 |
| 2 | CBX3 - Q13185 |
| 3 | ZNF10 - P21506 |
| 4 | ZNF268 - Q14587 |
| 5 | CBX1 - P83916 |
| 6 | CHAF1A - Q13111 |
| 7 | ZNF354A - O60765 |
| 8 | ESR1 - P03372 |
| 9 | ZNF382 - Q96SR6 |
| 10 | ZNF74 - Q16587 |
| 11 | TRIM24 - O15164 |
| 12 | NR3C1 - P04150 |
| 13 | VHL - P40337 |
| 14 | ZNF197 - O14709 |
| 15 | SETDB1 - Q15047 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Disease Linkage
Comments:
TIF1b contributes to EMT via E-cadherin and N-cadherin regulation in lung cancer cell lines.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +107, p<0.0008); Breast epithelial cell carcinomas (%CFC= +64, p<0.015); Cervical cancer stage 2B (%CFC= +72); Classical Hodgkin lymphomas (%CFC= +50, p<0.0006); Colon mucosal cell adenomas (%CFC= +81, p<0.0001); Gastric cancer (%CFC= +49, p<0.0002); Large B-cell lymphomas (%CFC= +127, p<0.005); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +181, p<0.0002); Ovary adenocarcinomas (%CFC= +95, p<0.032); Pituitary adenomas (ACTH-secreting) (%CFC= +105); Skin melanomas - malignant (%CFC= +195, p<0.0001); and Uterine fibroids (%CFC= +46, p<0.016). The COSMIC website notes an up-regulated expression score for TIF1b in diverse human cancers of 718, which is 1.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 17 for this protein kinase in human cancers was 0.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 25243 diverse cancer specimens. This rate is only -36 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.2 % in 603 endometrium cancers tested; 0.2 % in 589 stomach cancers tested; 0.18 % in 864 skin cancers tested; 0.16 % in 1270 large intestine cancers tested; 0.09 % in 273 cervix cancers tested; 0.09 % in 128 biliary tract cancers tested; 0.08 % in 1768 lung cancers tested; 0.08 % in 1512 liver cancers tested; 0.07 % in 548 urinary tract cancers tested; 0.05 % in 710 oesophagus cancers tested; 0.05 % in 1316 breast cancers tested; 0.04 % in 1360 kidney cancers tested; 0.03 % in 891 ovary cancers tested; 0.02 % in 558 thyroid cancers tested; 0.02 % in 1469 pancreas cancers tested; 0.01 % in 942 upper aerodigestive tract cancers tested; 0.01 % in 881 prostate cancers tested; 0.01 % in 2082 central nervous system cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,526 cancer specimens
Comments:
Only 1 deletion and 1 insertion and no complex mutations are noted on the COSMIC website.

