Nomenclature
Short Name:
LATS2
Full Name:
Serine-threonine protein kinase LATS2
Alias:
- EC 2.7.11.1
- FLJ13161
- KPM
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
NDR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
120136
# Amino Acids:
1088
# mRNA Isoforms:
1
mRNA Isoforms:
120,136 Da (1088 AA; Q9NRM7)
4D Structure:
Interacts with and is phosphorylated by STK6. Binds to AR. Interacts with JUB during mitosis and this complex regulates organization of the spindle apparatus through recruitment of gamma-tubulin to the centrosome. Interacts (via PPxY motif) with YAP1 (via WW domains). Interacts with MOBKL1A and MOBKL1B.
1D Structure:
Subfamily Alignment
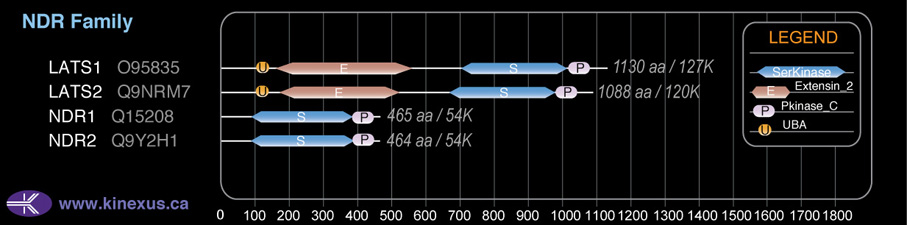
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 98 | 139 | UBA |
| 163 | 519 | Extensin_2 |
| 668 | 973 | Pkinase |
| 974 | 1052 | Pkinase_C |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K55, K565, K570, K571.
Serine phosphorylated:
S12, S83, S172, S288, S380, S408, S446, S528, S535, S543, S576, S598, S835, S872+, S1027, .
Threonine phosphorylated:
T5, T9, T10, T279, T690, T1024, T1026, T1041, .
Tyrosine phosphorylated:
Y11, Y82, Y135, Y286, Y531, Y1038, Y1051.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 48
48
1097
27
713
 44
44
999
9
1561
 -
-
-
-
-
 17
17
385
111
1567
 37
37
837
43
693
 5
5
110
36
119
 2
2
44
43
44
 21
21
484
18
1010
 1
1
22
3
2
 16
16
364
107
462
 3
3
75
11
95
 42
42
949
46
813
 57
57
1300
2
154
 21
21
471
5
652
 11
11
255
11
487
 16
16
355
22
383
 28
28
636
108
3336
 21
21
476
5
784
 16
16
354
77
1019
 43
43
974
117
757
 15
15
345
11
708
 3
3
59
9
43
 -
-
-
-
-
 69
69
1575
7
1408
 5
5
116
11
226
 43
43
990
77
830
 19
19
428
5
611
 34
34
769
5
1030
 78
78
1772
5
2315
 5
5
120
56
141
 22
22
507
12
66
 100
100
2281
28
4775
 15
15
349
81
1104
 48
48
1099
104
883
 31
31
704
61
991
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.5
99.5
99.7
99 70.6
70.6
71.2
- -
-
-
85 -
-
-
- 86.7
86.7
89.9
90 -
-
-
- 82.5
82.5
86.6
86 82.9
82.9
87
86 -
-
-
- 48.8
48.8
61
- 75.7
75.7
83.2
80 47.9
47.9
58.8
74 63.5
63.5
73
69 -
-
-
- 22.5
22.5
31.4
- 40.8
40.8
52.8
- -
-
-
48 38.6
38.6
52
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 28.1
28.1
42.1
- 24.5
24.5
36.4
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | JUB - Q96IF1 |
| 2 | SAV1 - Q9H4B6 |
| 3 | YAP1 - P46937 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 14-3-3-g | P61981 | S59 | VVGARRSSWRVISSI | ? |
| Snail1 | O95863 | T203 | QGHVRTHTGEKPFSC | ? |
| TAZ | Q16635 | S66 | GSHSRQSSTDSSGGH | |
| TAZ | Q16635 | S89 | AQHVRSHSSPASLQL | - |
| YAP1 | P46937 | S109 | KSHSRQASTDAGTAG | |
| YAP1 | P46937 | S127 | PQHVRAHSSPASLQL | - |
| YAP1 | P46937 | S164 | AQHLRQSSFEIPDDV | |
| YAP1 | P46937 | S381 | LRTMTTNSSDPFLNS | |
| YAP1 | P46937 | S61 | IVHVRGDSETDLEAL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
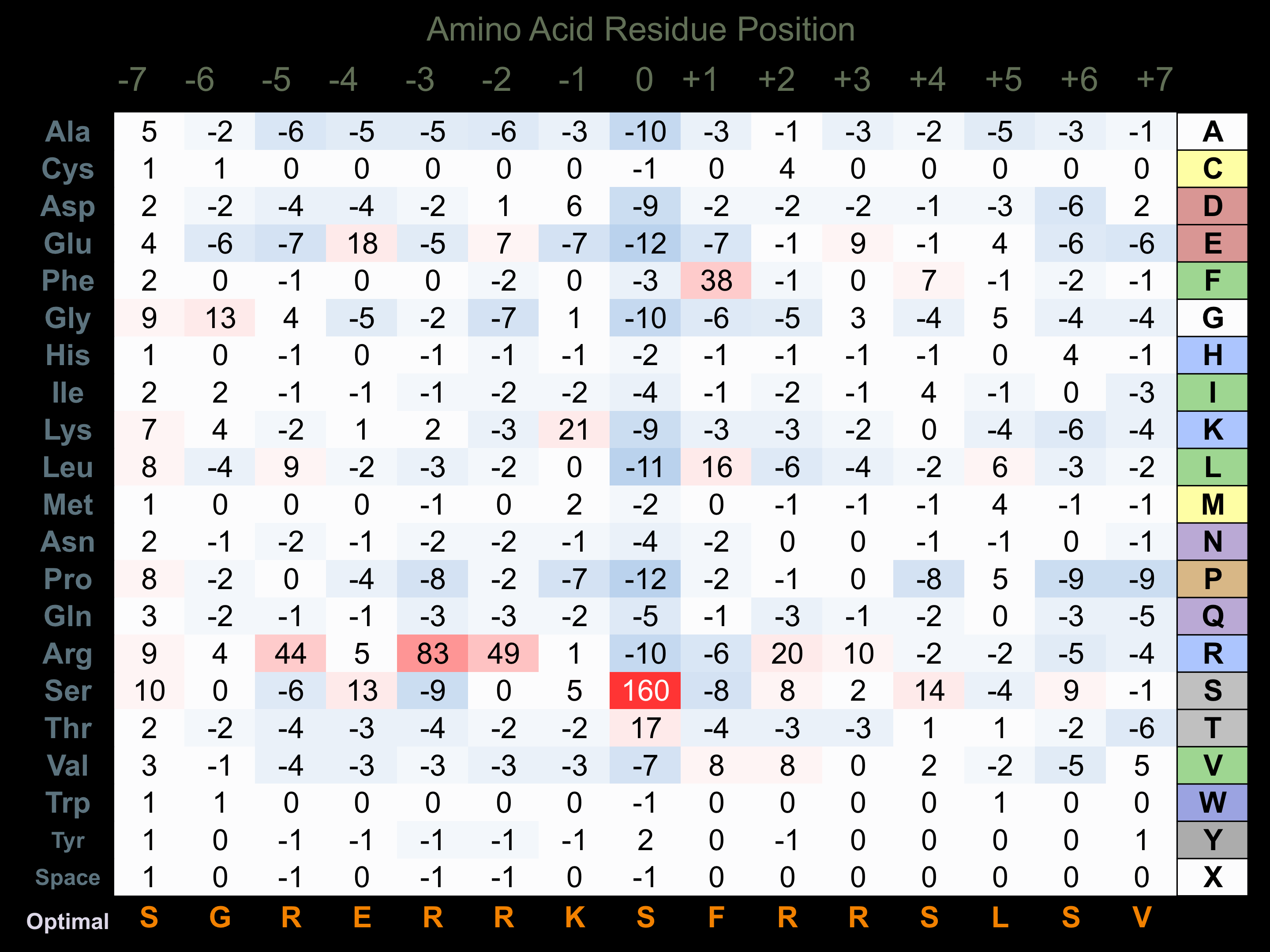
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Lestaurtinib | Kd = 1 nM | 126565 | 22037378 | |
| Staurosporine | Kd = 17 nM | 5279 | 18183025 | |
| GSK690693 | Kd = 140 nM | 16725726 | 494089 | 22037378 |
| Nintedanib | Kd = 380 nM | 9809715 | 502835 | 22037378 |
| SU14813 | Kd = 430 nM | 10138259 | 1721885 | 18183025 |
| Sunitinib | Kd = 460 nM | 5329102 | 535 | 18183025 |
| Dovitinib | Kd = 530 nM | 57336746 | 18183025 | |
| A674563 | Kd = 620 nM | 11314340 | 379218 | 22037378 |
| Ruboxistaurin | Kd = 640 nM | 153999 | 91829 | 18183025 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| KW2449 | Kd = 1.5 µM | 11427553 | 1908397 | 22037378 |
| Linifanib | Kd = 1.7 µM | 11485656 | 223360 | 18183025 |
| SB202190 | Kd = 1.7 µM | 5353940 | 278041 | 18183025 |
| NVP-TAE684 | Kd = 2 µM | 16038120 | 509032 | 22037378 |
| JNJ-28312141 | Kd = 2.1 µM | 22037378 | ||
| N-Benzoylstaurosporine | Kd = 2.2 µM | 56603681 | 608533 | 18183025 |
| AST-487 | Kd = 3 µM | 11409972 | 574738 | 18183025 |
| BMS-690514 | Kd < 3 µM | 11349170 | 21531814 | |
| PP242 | Kd = 4 µM | 25243800 | 22037378 | |
| R406 | Kd = 4 µM | 11984591 | 22037378 |
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Intracranial abscess
Comments:
LATS2 appears to be a tumour suppressor protein (TSP). Cancer-related mutations in human tumours point to a loss of function of the protein kinase. The active form of the protein kinase normally acts to inhibit tumour cell proliferation. K697A and S872A mutations lead to loss of tumour suppressor activity.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -73, p<0.081); Colorectal adenocarcinomas (early onset) (%CFC= +108, p<0.007); Large B-cell lymphomas (%CFC= +55, p<0.0006); and Ovary adenocarcinomas (%CFC= -54, p<0.038). The COSMIC website notes an up-regulated expression score for LATS2 in diverse human cancers of 304, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 33 for this protein kinase in human cancers was 0.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. S83E and S83C mutations in LATS2 can lead to failure to localize at the centromere during interphase. LATS2 deletion in mice was lethal at embryonic states, which indicates that it may have a critical role in maintainance of mitotic fidelity and genomic integrity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25371 diverse cancer specimens. This rate is only -6 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.42 % in 1093 large intestine cancers tested; 0.25 % in 589 stomach cancers tested; 0.23 % in 805 skin cancers tested; 0.2 % in 602 endometrium cancers tested; 0.09 % in 1941 lung cancers tested; 0.08 % in 1226 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A324V (18).
Comments:
Only 6 deletions, 1 insertion and no complex mutations are noted on the COSMIC website.

