Nomenclature
Short Name:
AurA
Full Name:
Serine-threonine-protein kinase 6
Alias:
- AIK
- Aurora/IPL1-related kinase 1
- Aurora-1
- Aurora-A
- Aurora-family kinase 1
- STK7
- ARK1
- AURKA
- Aurora kinase A
- Aurora,IPL1-related kinase 1
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
AUR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
45,809
# Amino Acids:
403
# mRNA Isoforms:
1
mRNA Isoforms:
45,809 Da (403 AA; O14965)
4D Structure:
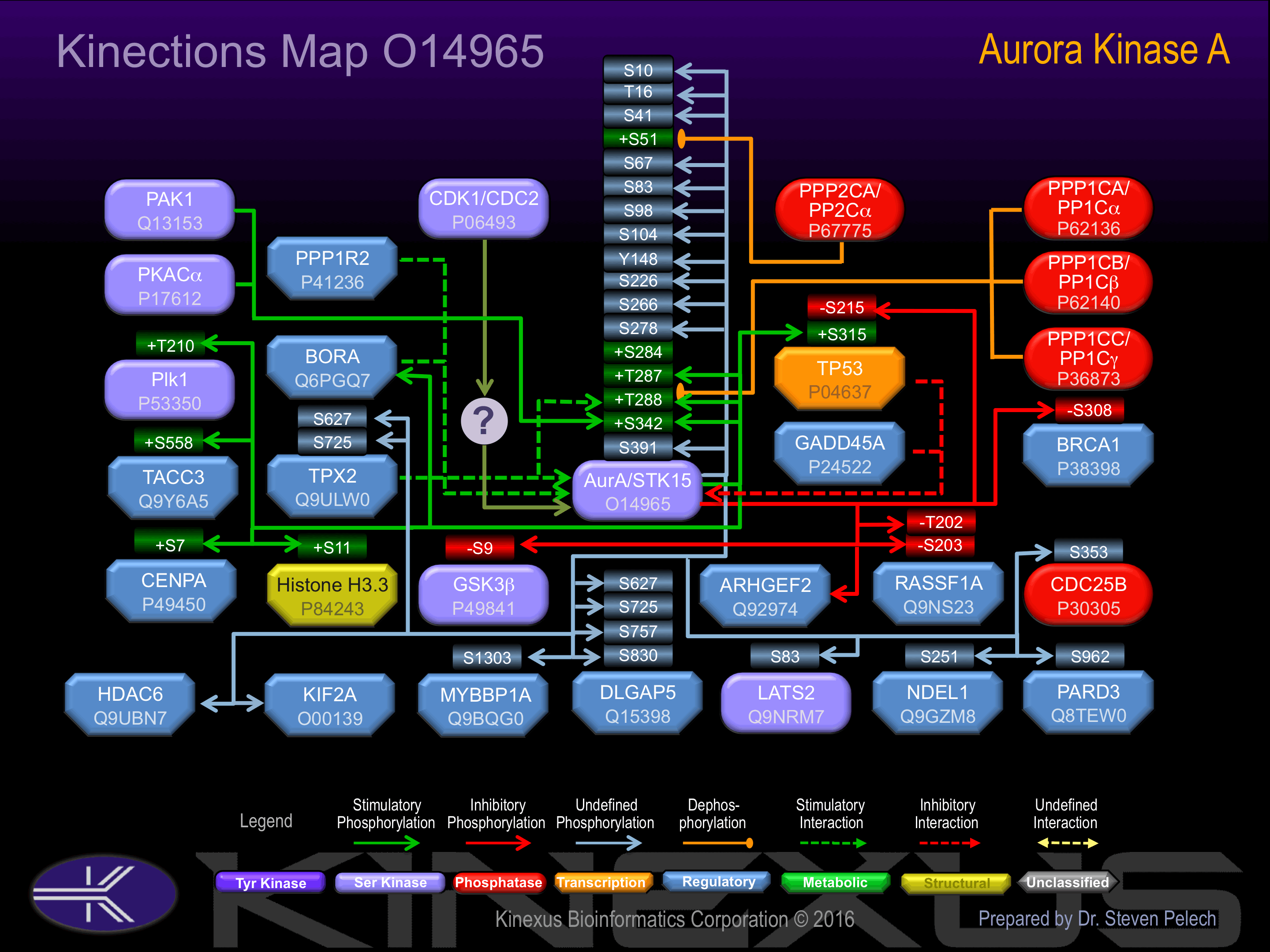
Interacts with TPX2. Interacts with TACC1 and CPEB1. Interacts with its substrates BORA, BRCA1, KIF2A, PARD3 and ARHGEF2. Interaction with BORA promotes phosphorylation of PLK1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 133 | 383 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S4, S10, S41, S51+, S53, S54, S66, S67, S83, S89, S98, S104, S116, S226, S266, S278, S284+, S342+, S369, S391.
Threonine phosphorylated:
T16, T287+, T288+, T292.
Tyrosine phosphorylated:
Y148, Y197, Y199, Y212, Y295.
Ubiquitinated:
K143.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 33
33
1192
42
2260
 0.6
0.6
22
22
14
 5
5
191
11
320
 31
31
1115
127
3766
 11
11
404
36
348
 9
9
315
119
1019
 8
8
304
47
555
 100
100
3610
55
7734
 13
13
468
24
361
 1.4
1.4
52
117
77
 2
2
81
38
143
 16
16
576
239
566
 2
2
71
44
170
 0.9
0.9
33
17
17
 2
2
68
28
105
 0.7
0.7
26
23
25
 2
2
81
214
633
 2
2
89
20
128
 2
2
69
125
113
 10
10
363
162
402
 3
3
96
28
244
 3
3
92
34
192
 5
5
190
14
226
 16
16
568
24
728
 7
7
259
28
409
 87
87
3133
79
8030
 3
3
123
47
173
 3
3
98
22
211
 2
2
78
24
111
 2
2
88
42
53
 12
12
441
30
530
 16
16
566
51
630
 3
3
102
86
456
 26
26
941
78
789
 21
21
741
48
851
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 100
100
100
100 47.8
47.8
62.2
97 -
-
-
89 -
-
-
85 89.9
89.9
93.1
90 -
-
-
- 82.6
82.6
86.8
84 83.6
83.6
87.3
85 -
-
-
- 65.5
65.5
70.2
- 73.1
73.1
82.9
75 65.9
65.9
77.2
67 53.4
53.4
65.8
63 -
-
-
- 38.5
38.5
57.8
53 53.1
53.1
67
- 46.6
46.6
56.3
62 50.6
50.6
60
- -
-
-
- -
-
-
- -
-
-
65 46.4
46.4
55.3
63 39
39
57.6
43 -
-
-
55
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TACC1 - O75410 |
| 2 | AURKAIP1 - Q9NWT8 |
| 3 | BRCA1 - P38398 |
| 4 | MBD3 - O95983 |
| 5 | CDC20 - Q12834 |
| 6 | PPP1CC - P36873 |
| 7 | NME1 - P15531 |
| 8 | PPP1CA - P62136 |
| 9 | UBE2N - P61088 |
| 10 | CHFR - Q96EP1 |
| 11 | PML - P29590 |
| 12 | CDC25B - P30305 |
| 13 | BORA - Q6PGQ7 |
| 14 | NIN - Q8N4C6 |
| 15 | PRKACA - P17612 |
Regulation
Activation:
Activated by phosphorylation at Ser-284, Thr-287, Thr-288 and Ser-342.
Inhibition:
NA
Synthesis:
Expression is cell-cycle regulated, low in G1/S, accumulates during G2/M, and decreases rapidly after.
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AurA | O14965 | S10 | RSKENCISGPVKATA | |
| AurA | O14965 | T16 | ISGPVKATAPVGGPK | |
| AurA | O14965 | S41 | QNPLPVNSGQAQRVL | |
| AurA | O14965 | S67 | QAQKLVSSHKPVQNQ | |
| AurA | O14965 | S83 | QKQLQATSVPHPVSR | |
| AurA | O14965 | S98 | PLNNTQKSKQPLPSA | |
| AurA | O14965 | S104 | KSKQPLPSAPENNPE | |
| AurA | O14965 | S226 | YRELQKLSKFDEQRT | |
| AurA | O14965 | S266 | PENLLLGSAGELKIA | |
| AurA | O14965 | S278 | KIADFGWSVHAPSSR | |
| AurA | O14965 | T287 | HAPSSRRTTLCGTLD | + |
| PKACa | P17612 | T288 | APSSRRTTLCGTLDY | + |
| AurA | O14965 | T288 | APSSRRTTLCGTLDY | + |
| PAK1 | Q13153 | T288 | APSSRRTTLCGTLDY | + |
| AurA | O14965 | S342 | QETYKRISRVEFTFP | + |
| PAK1 | Q13153 | S342 | QETYKRISRVEFTFP | + |
| AurA | O14965 | S391 | TANSSKPSNCQNKES |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Pin1 | Q13526 | S16 | PGWEKRMSRSSGRVY | - |
| AurA (STK15) | O14965 | S10 | RSKENCISGPVKATA | |
| AurA (STK15) | O14965 | S104 | KSKQPLPSAPENNPE | |
| AurA (STK15) | O14965 | S226 | YRELQKLSKFDEQRT | |
| AurA (STK15) | O14965 | S266 | PENLLLGSAGELKIA | |
| AurA (STK15) | O14965 | S278 | KIADFGWSVHAPSSR | |
| AurA (STK15) | O14965 | S342 | QETYKRISRVEFTFP | + |
| AurA (STK15) | O14965 | S391 | TANSSKPSNCQNKES | |
| AurA (STK15) | O14965 | S41 | QNPLPVNSGQAQRVL | |
| AurA (STK15) | O14965 | S67 | QAQKLVSSHKPVQNQ | |
| AurA (STK15) | O14965 | S83 | QKQLQATSVPHPVSR | |
| AurA (STK15) | O14965 | S98 | PLNNTQKSKQPLPSA | |
| AurA (STK15) | O14965 | T16 | ISGPVKATAPVGGPK | |
| AurA (STK15) | O14965 | T287 | HAPSSRRTTLCGTLD | + |
| AurA (STK15) | O14965 | T288 | APSSRRTTLCGTLDY | + |
| AurA (STK15) | O14965 | Y148 | KGKFGNVYLAREKQS | |
| BRCA1 | P38398 | S308 | KAEFCNKSKQPGLAR | |
| Cdc25B | P30305 | S353 | VQNKRRRSVTPPEEQ | |
| CENPA | P49450 | S7 | _MGPRRRSRKPEAPR | + |
| CPEB | Q9BZB8 | T172 | VRGSRLDTRPILDSR | |
| DLG7 | Q15398 | S627 | VKLFSGLSVSSEGPS | |
| DLG7 | Q15398 | S725 | CLSSERMSLPLLAGG | |
| DLG7 | Q15398 | S757 | EGMELNSSITSQDVL | |
| DLG7 | Q15398 | S830 | QEHARHISFGGNLIT | |
| FAF1 | Q9UNN5 | S289 | ITDVHMVSDSDGDDF | |
| FAF1 | Q9UNN5 | S291 | DVHMVSDSDGDDFED | |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| KIF2C | Q99661 | S192 | VNSVRRKSCLVKEVE | |
| KIF2C | Q99661 | S715 | MQLEEQASRQISSKK | |
| LATS2 | Q9NRM7 | S83 | ALREIRYSLLPFANE | ? |
| MYBBP1A | Q9BQG0 | S1303 | ARKKARLSLVIRSPS | |
| NDEL1 | Q9GZM8 | S251 | LTPSARISALNIVGD | |
| p53 | P04637 | S215 | DRNTFRHSVVVPYEP | - |
| p53 | P04637 | S315 | LPNNTSSSPQPKKKP | + |
| Plk1 (PLK) | P53350 | T210 | YDGERKKTLCGTPNY | + |
| RASSF1 | Q9NS23 | S207 | TSVRRRTSFYLPKDA | |
| TACC3 | Q9Y6A5 | S558 | ESALRKQSLYLKFDP | |
| TPX2 | Q9ULW0 | S121 | PAQPQRRSLRLSAQK | |
| TPX2 | Q9ULW0 | S125 | QRRSLRLSAQKDLEQ | |
| Vimentin | P08670 | S72 | SSAVRLRSSVPGVRL |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
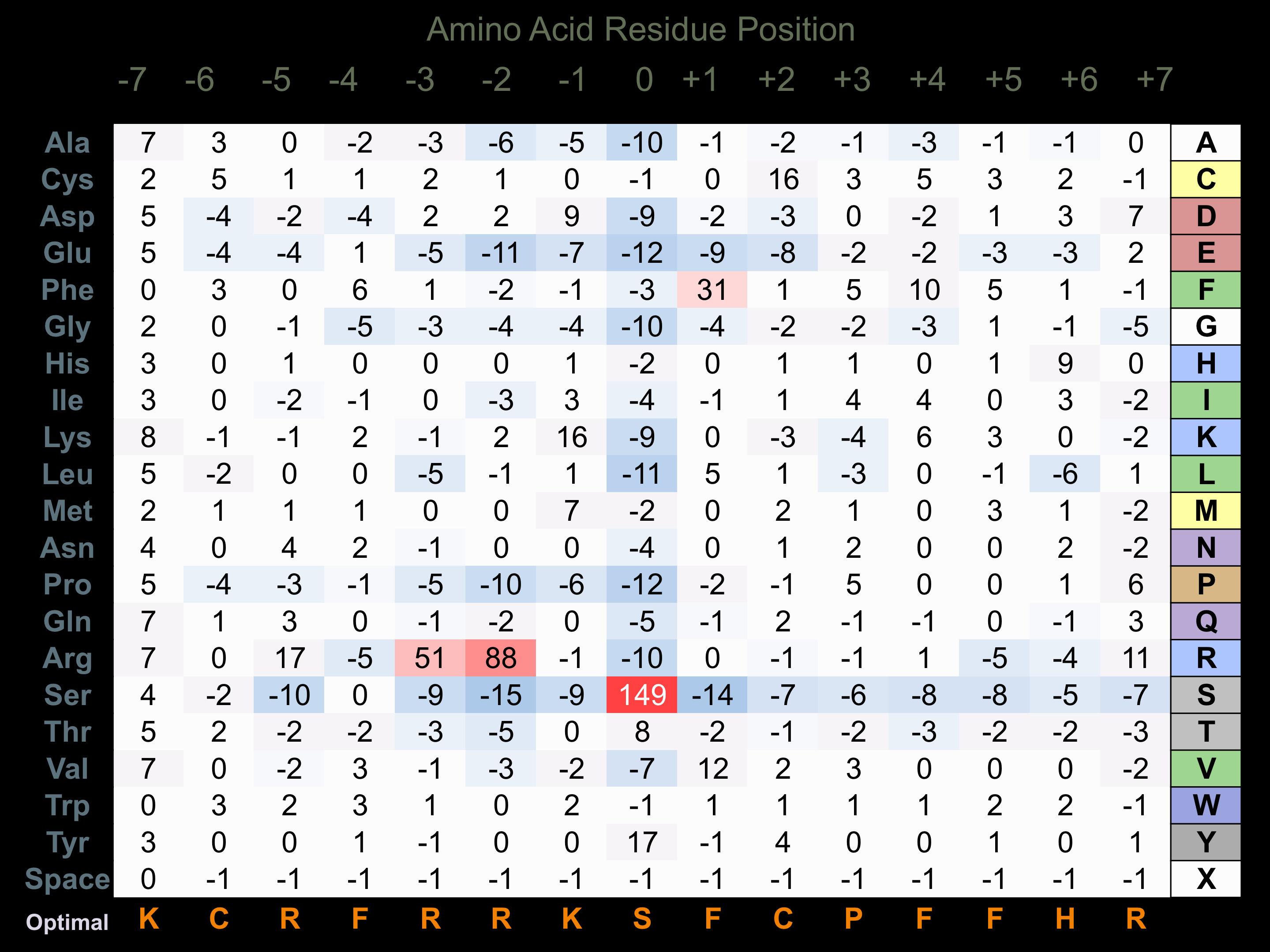
Matrix Type:
Experimentally derived from alignment of 62 known protein substrate phosphosites and 66 peptides phosphorylated by recombinant AurA in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Laryngeal carcinomas; Laryngeal squamous cell carcinomas; Breast cancer; Colorectal cancer; neuroblastomas; Colon cancer; Atypical teratoid rhabdoid tumours; neuroblastomas, susceptibility
Comments:
AurA appears to be a tumour requiring protein (TRP). The active form of the protein kinase normally acts to promote tumour cell proliferation. Although AurA is not an oncoprotein, it required for upstream oncoproteins to promote tumour formation. its low rate of mutation and down-regulation in human cancers supports it identification as a tumour-requiring protein, and as a target for cancer drug development.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +366, p<0.005); Breast non-basal-like cancer (BLC) (%CFC= +65, p<0.0001); Breast sporadic basal-like cancer (BLC) (%CFC= +81, p<0.0001); Cervical cancer (%CFC= -76, p<0.0001); Classical Hodgkin lymphomas (%CFC= +157, p<0.0001); Colon mucosal cell adenomas (%CFC= +121, p<0.0001); Head and neck squamous cell carcinomas (HNSCC) (%CFC= +47, p<0.003); Large B-cell lymphomas (%CFC= +224, p<0.0001); Lung adenocarcinomas (%CFC= +237, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +246, p<0.0001); Ovary adenocarcinomas (%CFC= +935, p<0.005); Prostate cancer - metastatic (%CFC= +90, p<0.0001); Prostate cancer - primary (%CFC= +87, p<0.039); Skin fibrosarcomas (%CFC= +471, p<0.0008); Skin melanomas - malignant (%CFC= +143, p<0.0001); Skin squamous cell carcinomas (%CFC= +183, p<0.008); and Vulvar intraepithelial neoplasia (%CFC= +411, p<0.0002). The COSMIC website notes an up-regulated expression score for AurA in diverse human cancers of 1011, which is 2.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 6 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. AurA phosphotransferase activity can be abrogated with a K162R mutation, decreased 10-fold with G198N (which also induces abnormal binding to INCENP and BIRC5), and increased with T288D. Phosphatase type I interactions can be decreased with F165A, and F346A mutations, while ubiquitination can be reduced with R205A. Autophosphorylation can be inhibited with D274N, and TPX2 interactions can be interrupted with a T287E mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25634 diverse cancer specimens. This rate is very similar (+ 3% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.54 % in 1184 large intestine cancers tested; 0.34 % in 805 skin cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,852 cancer specimens
Comments:
Only 1 deletion, no insertions and no complex mutations are noted on the COSMIC website.