Nomenclature
Short Name:
ErbB2
Full Name:
Receptor tyrosine-protein kinase erbB-2
Alias:
- CD340
- HER-2
- Kinase ErbB2
- MLN 19
- V-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
- C-erbB-2
- EC 2.7.10.1
- Epidermal growth factor receptor-related protein
- ErbB2
- HER2
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
EGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
137,910
# Amino Acids:
1255
# mRNA Isoforms:
6
mRNA Isoforms:
137,910 Da (1255 AA; P04626); 136,501 Da (1240 AA; P04626-4); 134,856 Da (1225 AA; P04626-5); 97,382 Da (888 AA; P04626-6); 70,917 Da (645 AA; P04626-2); 62,568 Da (569 AA; P04626-3)
4D Structure:
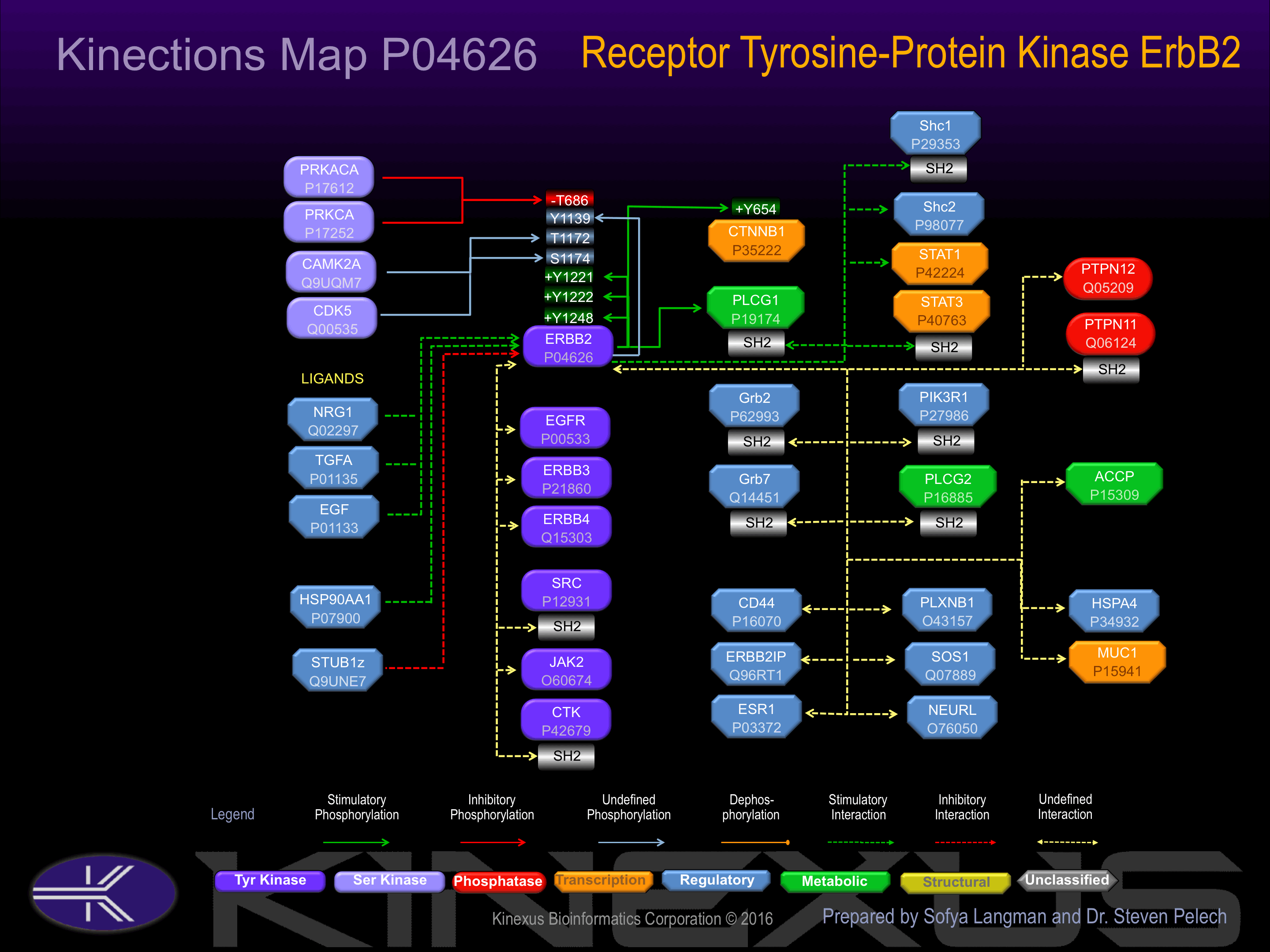
Heterodimer with each of the other ERBB receptors Potential. Interacts with PRKCABP and PLXNB1. Part of a complex with EGFR and either PIK3C2A or PIK3C2B. May interact with PIK3C2B when phosphorylated on Tyr-1196. Interacts with MEMO when phosphorylated on Tyr-1248. Interacts with MUC1. Stimulation by heregulin (HRG) in breast cancer cell lines induces binding of MUC1 with gamma-catenin.
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
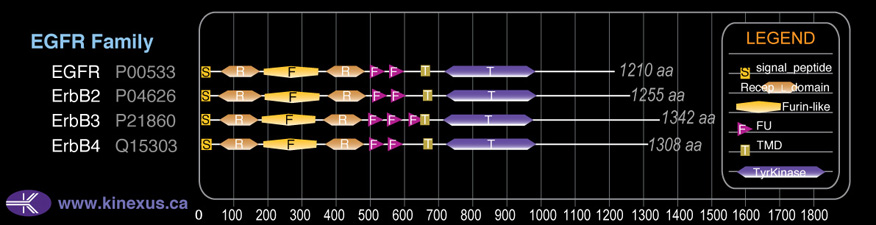
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 22 | signal_peptide |
| 52 | 173 | Recep_L_domain |
| 189 | 343 | Furin-like |
| 366 | 486 | Recep_L_domain |
| 501 | 552 | FU |
| 557 | 606 | FU |
| 653 | 675 | TMD |
| 720 | 976 | TyrKc |
| 720 | 978 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N68, N124, N187, N259,N530, N629.
Serine phosphorylated:
S703, S728, S998, S1002, S1007, S1049, S1050, S1051, S1054, S1066, S1083, S1107, S1122, S1134, S1151, S1174, S1214, S1235.
Threonine phosphorylated:
T182, T686-, T701, T733, T875, T948, T1003, T1052, T1103, T1124, T1132, T1166, T1172, T1198, T1236, T1240, T1242.
Tyrosine phosphorylated:
Y735, Y772, Y877+, Y923, Y1005, Y1023, Y1112-, Y1127, Y1139, Y1196+, Y1221+, Y1222+, Y1248+.
Ubiquitinated:
K150, K175, K724, K736, K747, K753, K860, K887.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 91
91
927
38
1255
 5
5
52
16
59
 25
25
253
28
365
 55
55
556
144
1417
 75
75
762
35
706
 9
9
90
89
114
 49
49
493
49
622
 71
71
723
75
1299
 69
69
699
17
606
 23
23
238
125
235
 15
15
155
52
234
 58
58
589
231
582
 16
16
159
50
327
 11
11
113
17
107
 30
30
307
44
617
 3
3
27
21
32
 30
30
305
320
2110
 25
25
257
40
293
 13
13
130
133
239
 52
52
526
137
574
 19
19
192
46
266
 13
13
132
49
218
 19
19
191
46
227
 21
21
212
39
473
 18
18
181
46
332
 81
81
825
104
1863
 20
20
205
53
224
 35
35
354
41
430
 24
24
245
40
374
 7
7
67
42
46
 48
48
486
36
339
 100
100
1014
41
3344
 37
37
377
106
714
 54
54
550
88
543
 5
5
55
48
49
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 48
48
63.1
100 98.3
98.3
98.6
98 -
-
-
92 -
-
-
98 39.5
39.5
51.7
93 -
-
-
- 87.7
87.7
92
88 88
88
91.9
88 -
-
-
- 48
48
62.9
- 27.2
27.2
36.2
72 22.6
22.6
36.6
62.5 54.3
54.3
66.8
59 -
-
-
- 30.6
30.6
45.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | IGKC - P01834 |
| 2 | GRB2 - P62993 |
| 3 | SHC1 - P29353 |
| 4 | EGFR - P00533 |
| 5 | ERBB3 - P21860 |
| 6 | CTNNB1 - P35222 |
| 7 | PLCG1 - P19174 |
| 8 | PIK3R1 - P27986 |
| 9 | IGHG1 - P01857 |
| 10 | PIK3R2 - O00459 |
| 11 | ERBB4 - Q15303 |
| 12 | CD44 - P16070 |
| 13 | SRC - P12931 |
| 14 | MATK - P42679 |
| 15 | STAT3 - P40763 |
Regulation
Activation:
Phosphorylation of Tyr-1139 induces interaction with Grb2, Grb7 and Src. Phosphorylation of Tyr-1196 induces interaction with Crk. Phosphorylation of Tyr-1222 induces interaction with CGI-27, Grb2, and Shc1.
Inhibition:
Phosphorylation of Tyr-1112 induces interaction with Cbl, which promotes degradation.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CTNNB1 | P35222 | Y654 | RNEGVATYAAAVLFR | + |
| ErbB2 (HER2) | P04626 | Y1139 | TCSPQPEYVNQPDVR | ? |
| ErbB2 (HER2) | P04626 | Y1221 | SPAFDNLYYWDQDPP | + |
| ErbB2 (HER2) | P04626 | Y1222 | PAFDNLYYWDQDPPE | + |
| ErbB2 (HER2) | P04626 | Y1248 | PTAENPEYLGLDVPV | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
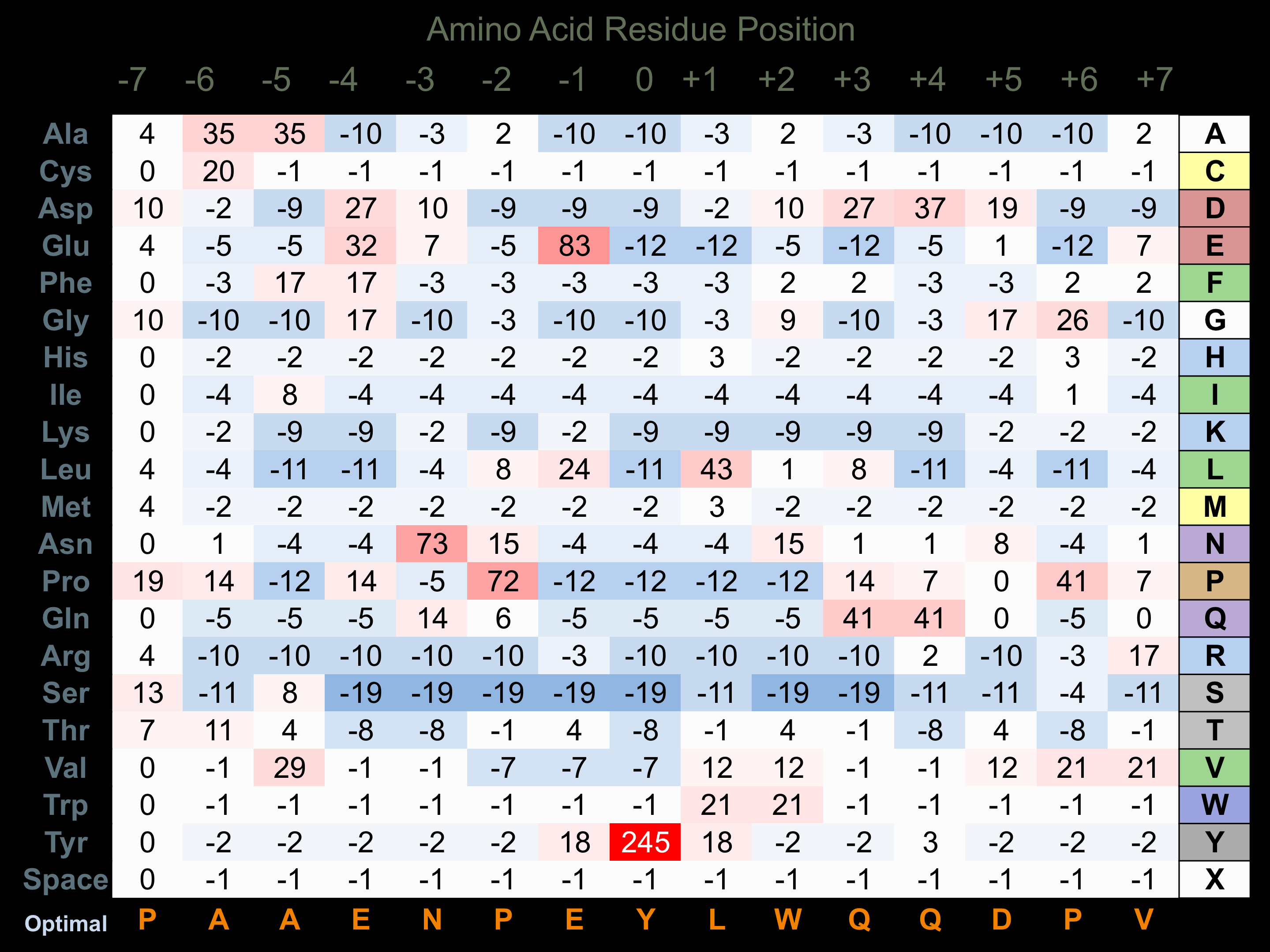
Matrix Type:
Experimentally derived from alignment of 23 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Wallerian degeneration; Charcot-Marie-Tooth disease Type 1a; Charcot-Marie-Tooth disease Type 1; Congenital pulmonary airway malformation
Comments:
Wallerian degeneration, also known as anterograde degeneration, is the degeneration of the distal segment of an axon after separation from the cell body by axotomy (transection of an axon). A similar process occurs in certain neurodegenerative diseases (termed 'Wallerian-like degeneration'), particularly those with components of impaired axonal transport. Charcot-Marie-Tooth disease is an inherited neurological disease affecting the peripheral nervous system. Symptoms include weakness, atrophy of the muscles, and loss of peripheral sensation. This disease is inherited in an autosomal dominant manner. It is thought that the symptoms of this disease are attributable to the progressive degeneration of axons. Nerve biopses from affected individuals reveal segments of demyelination, schwann cell hyperplasia, and evidence of attempted remyelination. Progression of the disease is characterized by progressive Schwann cell loss, indicating further demyelination and axonal degeneration. Overexpression of ErbB2 has been reported in schwann cells in nerve biopses from patients with Charcot-Marie-Tooth disease, hypothesized to have a role in the failure of myelination by schwann cells or contribute to the axonal damage observed in the disease. Therefore, elevated expression of ErbB2 may have a role in the pathogenesis of Charcot-Marie-Tooth disease.
Specific Cancer Types:
Breast cancer; Lung cancer (LC); Ductal carcinomas in situ; Uterine carcinosarcoma; Adenocarcinomas; Ovarian cancer; Medulloblastoma; Cholangiocarcinomas; Transitional cell carcinomas; Inflammatory breast carcinomas; Adenocarcinomas in situ; Pleomorphic adenoma; Primary peritoneal carcinomas; Sweat gland carcinomas; Placental site trophoblastic tumours; In situ carcinomas; Thymic epithelial neoplasm; Mammary Paget's disease; Hereditary diffuse gastric cancer; Breast fibroadenoma; Hidradenoma; Fallopian tube cancer; Diffuse gastric cancer; Pituitary carcinomas; Breast carcinomas in situ; Female breast carcinomas; Hidradenocarcinomas; Bile duct adenoma; Ovarian carcinosarcoma; Ewing's family of tumours; Microglandular adenosis; Sinonasal undifferentiated carcinomas; Nonseminomatous germ cell tumours; Papillary serous adenocarcinomas; Gastroesophageal junction adenocarcinomas; Glassy cell carcinomas of the cervix; Progesterone-receptor positive breast cancer; Lipid-rich carcinomas; Breast scirrhous carcinomas; neuroblastomas; Gastric cancer, somatic; Ovarian cancer, somatic; Adenocarcinomas of lung, somatic; Glioblastoma, somatic
Comments:
ERBB2 is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. Significantly elevated expression levels of ErbB2 is observed in many cancer types, including adenocarcinoma of the salivary gland, gastric cancer, ductal carcinoma, breast cancer, lung cancer, and ovarian cancer. Analysis of 120 primary lung tumours revealed that 4% had mutations in the ErbB2 catalytic domain, which was compared to a observed rate of 10% in lung adenocarcinoma cell lines. Several mutations in the ErbB2 gene have been observed in cancer patients, including a 4 amino-acid insertion at codon 774, a 3 amino-acid insertion at codon 779, and four substitution mutations (L755P, E914K, G776, N857S). However, gain-of-function mutations in ErbB2 that result in an over-active oncogenic protein are rarely seen in human cancers, insteadthe observed mutations are associated with an elevated expression of unmutated ErbB2. For example, ErbB2 expresion was observed to be ~20-30% higher in breast cancer cell lines compared to control breast tissue. For these cancer types, treatment with trastuzumab (Herceptin), an anti-ErbB2 antibody, has been demonstrated to be an effective anti-cancer treatment.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +190, p<0.024); Brain glioblastomas (%CFC= -56, p<0.031); Cervical cancer (%CFC= +88, p<0.079); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -76, p<0.003); Gastric cancer (%CFC= +237, p<0.101); Oral squamous cell carcinomas (OSCC) (%CFC= -50, p<0.039); Ovary adenocarcinomas (%CFC= +67, p<0.056); and Skin melanomas - malignant (%CFC= -55, p<0.078). The COSMIC website notes an up-regulated expression score for ERBB2 in diverse human cancers of 697, which is 1.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 42 for this protein kinase in human cancers was 0.7-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.11 % in 47546 diverse cancer specimens. This rate is a modest 1.4-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.58 % in 716 urinary tract cancers tested; 0.33 % in 2040 large intestine cancers tested; 0.25 % in 1275 stomach cancers tested; 0.18 % in 1426 skin cancers tested; 0.14 % in 752 endometrium cancers tested; 0.13 % in 3036 breast cancers tested; 0.11 % in 14526 lung cancers tested; 0.09 % in 1748 ovary cancers tested; 0.09 % in 1389 pancreas cancers tested; 0.06 % in 1709 upper aerodigestive tract cancers tested; 0.06 % in 1356 kidney cancers tested; 0.05 % in 2277 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A775ins (54); S310F (43); L755S (32); R678Q (18); (V842I (17); V777L (9); D776Y (6).
Comments:
Only 4 deletions over the full sequence, but 138 insertions (at A775) and 16 complex mutations (at G776) observed in the kinase domain are noted on the COSMIC website.